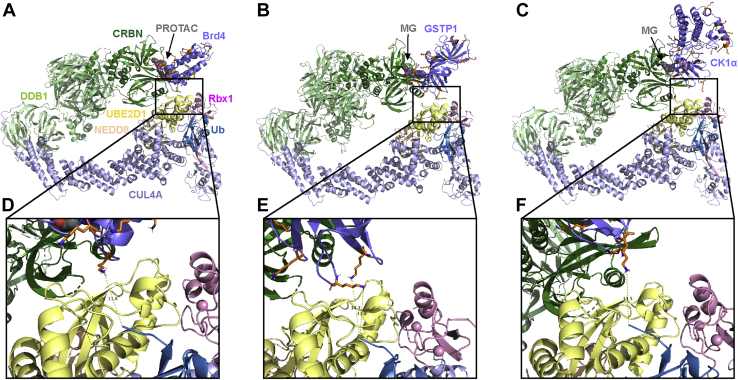
Figure 4.
Published ternary complex structures align with CRL4A ligase complex models.A and D, Brd4 – dBET6 (PROTAC) – CRBN (PDBID: 6BOY) aligned with CRL4A ligase complex conformation 2. Residue Lys141 of Brd4 is ∼11.8 Å to C-terminal of Ub. B and E, GSTP1 – CC-885 (molecular glue) – CRBN (PDBID: 5HXB) aligned with CRL4A ligase complex conformation 4. Residue Lys541 and Lys622 of GSTP1 are 11.8 Å and 14.3 Å to C-terminal of Ub, respectively. C and F, CK1α – lenalidomide (molecular glue) – CRBN (PDBID: 5FQD) aligned with CRL4A ligase complex conformation 2. Residue Lys51 of CK1α is 13.5 Å to C-terminal of Ub. Target proteins (Brd4, GSTP1, and CK1α) are shown in slate; Lys residues are shown in stick, orange; CRBN is shown in forest; DDB1 is shown in pale green; Cul4A is shown in light blue; Rbx1 is shown in pink; NEDD8 is shown in wheat; E2 (UBE2D1) is shown in pale yellow; ubiquitin is shown in sky blue and PROTAC and MGs are shown in sphere, gray.