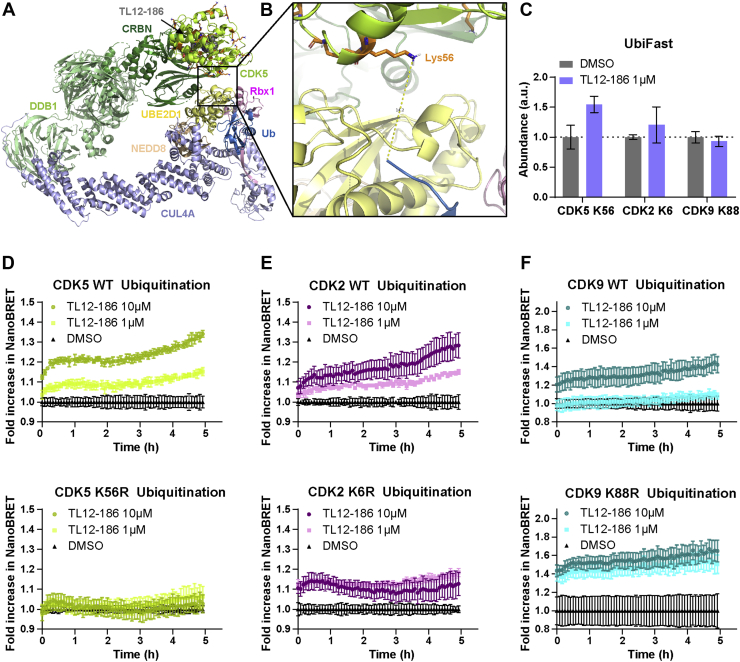
Figure 6.
Point mutant generation with predicted ubiquitin accessible lysine residues of CDK5, CDK2, and CDK9.A and B, CDK5 – CRL4A ligase complex conformation 2 model with Lys56 as the predicted ubiquitin accessible lysine residue. CDK5 is shown in lemon; Lys residues are shown in stick, orange; CRBN is shown in forest; DDB1 is shown in pale green; Cul4A is shown in light blue; Rbx1 is shown in pink; NEDD8 is shown in wheat; E2 (UBE2D1) is shown in pale yellow; ubiquitin is shown in sky blue; and TL12–186 is shown in sphere, gray. C, predicted ubiquitin accessible lysine residues ubiquitination level measured by UbiFast assay. Data are presented as mean ± SD of n = 2 technical replicates. D, CDK5, (E) CDK2, and (F) CDK9, both WT and mutant, kinetic ubiquitination measured by NanoBRET ubiquitination assay. Data are presented as mean ± SD of n = 4 technical replicates.