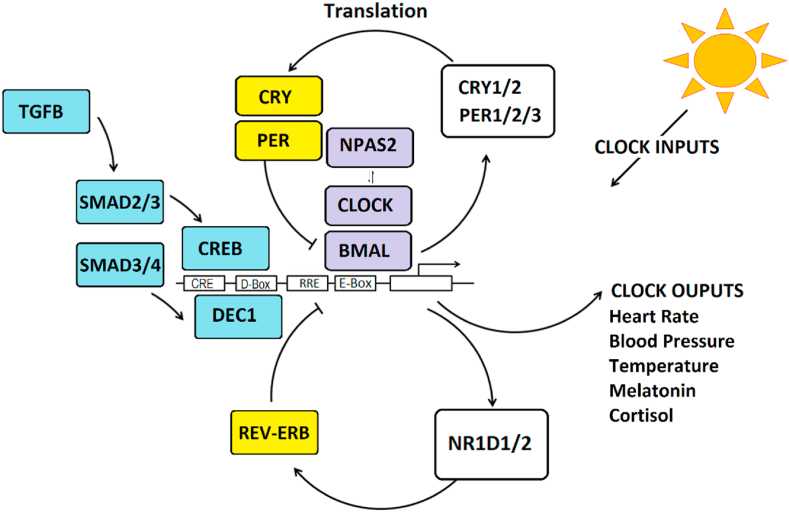
Fig. 1.
The molecular clock is comprised of clock genes, organized in transcriptional/translational feedback loops. Transcriptional activator proteins (purple), BMAL1 and CLOCK/NPAS2 bind to DNA elements in promoters (E-Box) to stimulate transcription of CRY1/2, PER1/2/3, and NR1D1/2 genes that encode transcriptional repressors (yellow) to regulate activity through protein and DNA interactions. Other regulators include DEC1 that binds to D-box elements. These interactions yield oscillatory changes in gene expression and translation over ∼24 h cycles causing cellular rhythms. Light and environmental entrainment signals can phase shift the clock through intracellular signals that stimulate the cAMP-response element binding protein (CREB) that binds to CRE DNA elements. Cytokine pathways (blue) directed by Transforming Growth Factor Beta (TGFB) and SMAD proteins can shift rhythms in response to changes in pH (through DEC1) and peripheral synchronization cues (through CREB). The circadian clock affects the expression of clock-controlled genes that are rhythmically expressed and control numerous physiological processes that require precise timing across the daily 24 h cycle. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)