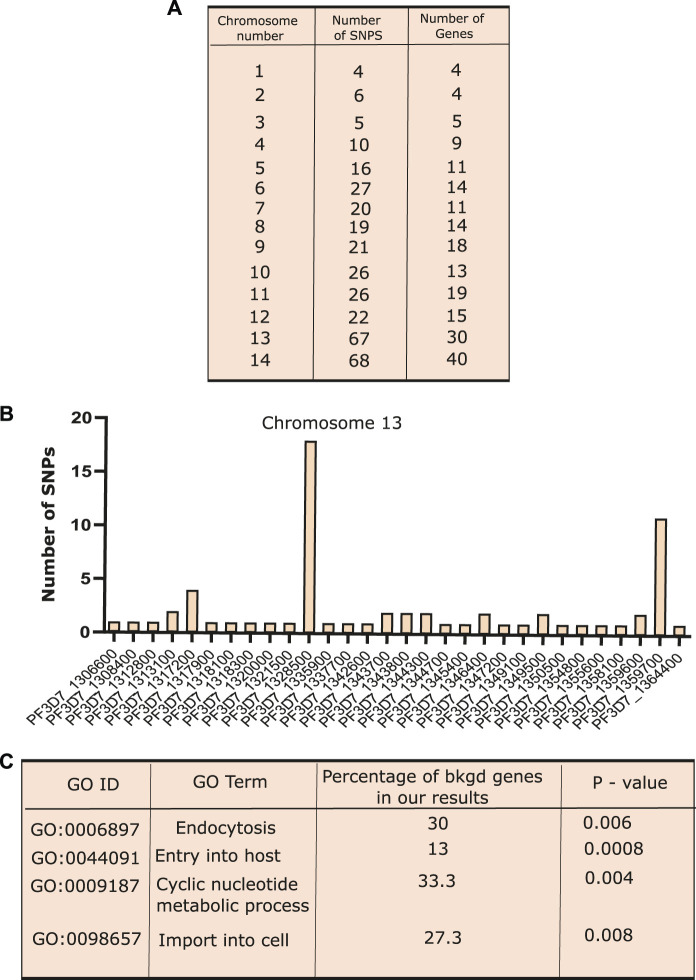
FIGURE 3.
Co-existence of different SNPs along with PfKelch13 mutations. (A) Exploration of background genomic variants co-existing with the K13 mutations in artemisinin resistance was identified using the 359 samples, which show one of the three prime K13 mutations (C580Y, R539T, and Y493H). In order for an SNP to be filtered for co-existence with K13 mutations, it had to be present in at least 75% of K13 mutant isolates. Variant annotation was done for all the SNPs using the tool snpEFF and only those SNPs that were non-synonymous mutation were considered for further analysis. Table presenting the number of SNPs identified present in the 75% of the K13 mutants isolates and present in less than 25% of the sensitive (K13 mutant absent) isolates in various chromosomes. (B) Bar graph showing the number of SNPs present on the different genes over chromosome 13. (C) Gene ontology performed for the genes showing SNP co-existing with K13 mutations. Different biological processes like “endocytosis”, “locomotion”, “cell division”, and “response to drug” were found to be enriched. Plots were generated using R and GraphPad.