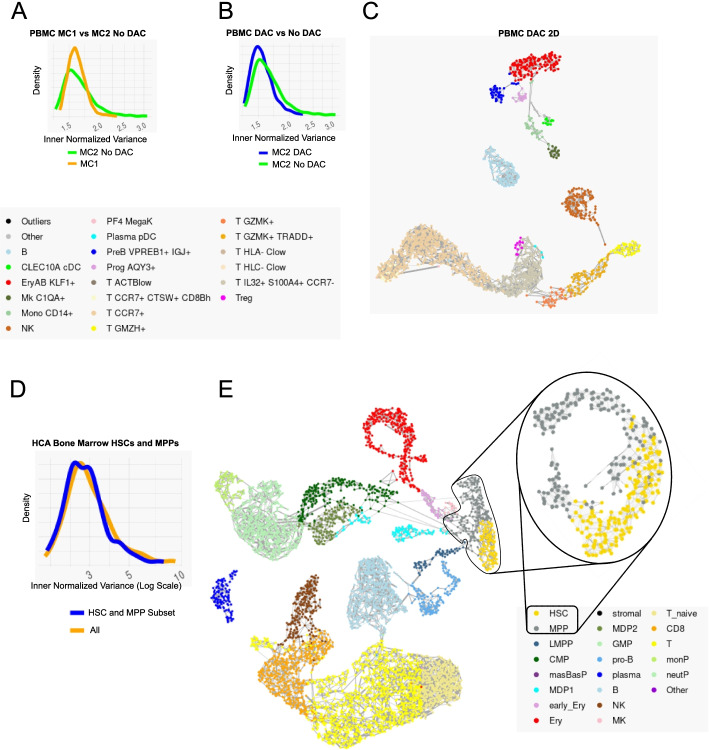
Fig. 2.
Robustness of the divide-and-conquer metacell algorithm. A Distribution of metacell normalized inner variance for the PBMC dataset, using the Baran et al. algorithm (orange) vs. MC2 two-sided stability score optimization, working on the entire data in a single pile (i.e., no divide and conquer, green). B Distribution of normalized inner variance for the PMBC dataset using the full MC2 algorithms (blue) vs. the single-pile algorithm (green). C Metacell graph derived by MC2 on the PMBC dataset. Annotation as in Baran et al. D Distribution of metacell normalized inner variance for HSC and MPP cells, when using full MC2 on the HCA bone marrow data set (orange) or when restricting analysis to MPP/HSC cells alone (blue). E Metacell graph for the full HCA BM data set and for metacells computed on the zoomed-in HSC/MPP subset