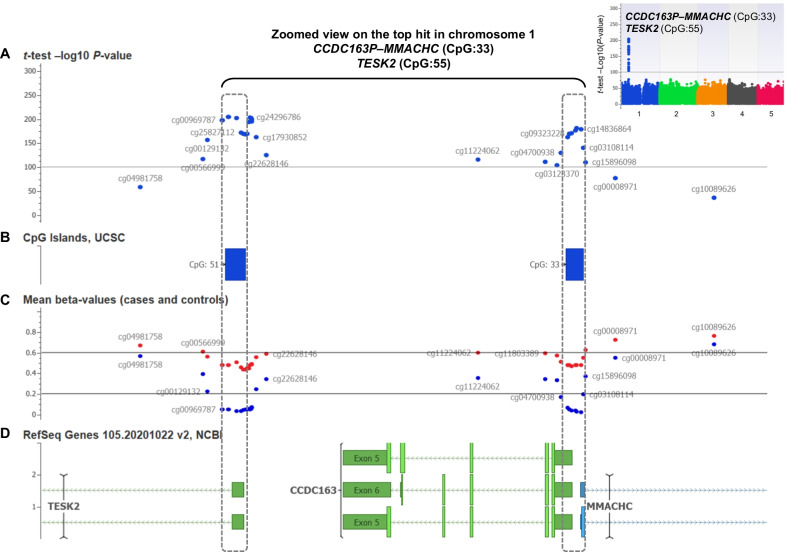
Fig. 3.
A Zoomed view of the epi-Manhattan plot reporting the epigenome-wide association study results that compared 17 patients with MMACHC epimutation (epi-cblC disease, isolated MMACHC epimutation, biallelic MMACHC epimutation) with controls. The − log10 P-value reports the t-test that compared the mean β values between the two groups. The horizontal line indicates a P-value threshold of 1 × 10−100. The zoomed view of the top epigenomic signature in chromosome 1 confirmed the significant association at the CpG island ‘CpG:33’ on the CCDC163–MMACHC bidirectional promoter but also revealed a second significant association at the CpG island ‘CpG:51’ on the promoter region of the TESK2 gene. B Positions of the CpG islands CpG:33 and CpG:51 according to the CpG Islands UCSC annotation. C Mean β values among the 17 patients with MMACHC epimutation (epi-cblC disease, isolated MMACHC epimutation, biallelic MMACHC epimutation) (red dots) and controls (blue dots). The horizontal lines correspond to β value thresholds of 0.2, below which the CpG probe is considered to be fully unmethylated. Above 0.6, the CpG probe is considered fully methylated. A β value between 0.2 and 0.6 indicates a hemimethylated CpG probe. D Genomic annotation according to RefSeq Genes 105.20201022 v2, NCBI