Abstract
Background:
We previously reported that the single nucleotide polymorphism (SNP) rs9282860 in serine threonine kinase 11 (STK11) gene which codes for liver kinase B1 (LKB1) has higher prevalence in White relapsing-remitting multiple sclerosis (RRMS) patients than controls. However it is not known if this SNP is a risk factor for MS in other populations.
Methods:
We assessed the prevalence of the STK11 SNP in samples collected from African American (AA) persons with MS (PwMS) and controls at multiple Veterans Affairs (VA) Medical Centers and from a network of academic MS centers. Genotyping was carried out using a specific Taqman assay. Comparisons of SNP frequencies were made using Fisher’s exact test to determine significance and odds ratios. Group means were compared by appropriate t-tests based on normality and variance using SPSS V27.
Results:
There were no significant differences in average age at first symptom onset, age at diagnosis, disease duration, or disease severity between RRMS patients recruited from VAMCs versus non-VAMCs. The SNP was more prevalent in AA than White PwMS, however only in secondary progressive MS (SPMS) patients was that difference statistically significant. AA SPMS patients had higher STK11 SNP prevalence than controls; and in that cohort the SNP was associated with older age at symptom onset and at diagnosis.
Conclusions:
The results suggest that the STK11 SNP represents a risk factor for SPMS in AA patients, and can influence both early (onset) and later (conversion to SPMSS) events.
Keywords: African American, Multiple sclerosis, SNP, Military Veterans, race and ethnicity, Progressive form
1. Introduction
Genetic studies have identified over 200 single nucleotide polymorphisms (SNPs) associated with an increased risk of multiple sclerosis (MS) (International_Multiple_Sclerosis_Genetics_Consortium 2019, International_Multiple_Sclerosis_Genetics_Consortium 2018, Baranzini and Oksenberg, 2017, Patsopoulos et al., 2011, Beecham et al., 2013); however, the majority of studies were done in persons of European ancestry. In contrast, few studies have examined the association of genetic variants with MS in African American (AA) populations, and of those most examined polymorphisms within major histocompatibility locus human leukocyte antigen (HLA) alleles (Isobe et al., 2013, McElroy et al., 2010, Johnson et al., 2010, Cree et al., 2009, Oksenberg et al., 2004). Outside the MHC region, several MS variants were shown to replicate in AA populations, although some studies (Beecham et al., 2020, Isobe et al., 2015) reported low replication pointing to significant heterogeneity amongst AA populations. Differences in genetic risk factors for MS could contribute to differences in MS incidence, prevalence or progression between AA and White populations (Amezcua and McCauley, 2020, Amezcua et al., 2018, Ventura et al., 2017, Cree et al., 2004).
We previously reported that a single nucleotide polymorphism (SNP) in the gene STK11 (serine-threonine kinase 11), which encodes LKB1 (liver kinase B1), is a risk factor in White RR (relapsing-remitting) patients with MS (PwMS), with an odds ratio (OR) of 1.63 in females (Boullerne et al., 2015). The SNP is present in intron 5 of the STK11 gene, and the C>T alteration removes a consensus CRE transcription factor binding site, which could influence STK11 gene expression. LKB1 is a ubiquitously expressed kinase, which activates multiple downstream kinases (Gan and Li, 2014) regulating cell functions including metabolism, migration, and proliferation. LKB1 is a metabolic sensor which helps maintain ATP levels during periods of intense activity and stress (Sebbagh et al., 2011). Roles for LKB1 in MS are suggested from several mouse showing that LKB1 depletion from spinal cord neurons led to axonal degeneration, macrophage infiltration, and hindlimb paralysis (Sun et al., 2011); mice with LKB1 depletion from regulatory Tcells developed an early onset autoimmune disease (Wu et al., 2017); and depletion from B-lymphocytes led to their spontaneous activation (Walsh et al., 2015). Conditional depletion of LKB1 from Schwann cells revealed a role in peripheral myelination (Beirowski, 2019, Pooya et al., 2014, Shen et al., 2014); while we showed that conditional depletion from astrocytes exacerbated disease in a mouse model of MS, associated with increased neuroinflammation and damage to spinal cord motor neurons (Kalinin et al., 2020). Alterations in LKB1 activity may therefore account for the STK11 SNP being a risk factor for MS.
Our previous study examined STK11 SNP prevalence in United States PwMS of European descent. To address whether this SNP is a risk factor in other populations, we examined its frequency in PwMS recruited at multiple Veterans Affairs Medical Centers (VAMCs) and compared that to its frequency in samples from PwMS and controls collected at academic institutes and a non-for-profit MS tissue repository. Previous epidemiological studies have suggested increased risk and greater disease severity of MS in military veterans compared to civilians (Wallin et al., 2000, Wallin et al., 2004, Wallin et al., 2012, Wallin et al., 2014, Wallin et al., 2018, Wallin et al., 2019); however, whether genetic factors contribute to those differences has not been addressed. Our findings show that the STK11 SNP is a risk factor for AA SPMS patients; and in contrast to White PwMS where the age at first symptom onset and diagnosis is younger in SNP carriers, in AA PwMS it is associated with older ages.
2. Methods and Materials
2.1. Study Population
The study population was comprised of 5 cohorts (Table 1). Plasma samples (n=91) were obtained from AA and White PwMS who participated in the VA Longitudinal MS Study (VALOMS) (Royal et al., 2012). Informed consent was obtained from subjects between the ages of 18 and 65 years at enrollment; with a diagnosis of definite MS based on McDonald Criteria (Polman et al., 2005, Polman et al., 2011). Genomic DNA (gDNA) samples (n=109) from AA PwMS were also obtained from the Accelerated Cure Project for Multiple Sclerosis (ACP) Repository (Waltham, MA). gDNA samples from AA controls were provided by Jorge Oksenberg, UCSF (n=398) and from ACP (n=42). Comparisons were made to previously described (Boullerne et al., 2015) cohorts of White PwMS (n=654) and controls (n=661). Patients in all cohorts were treated with interferon-beta or glatiramer acetate which are now known not to interfere with long-term progression or conversion to SPMS. All procedures were approved by the Baltimore VAMC Research and Development Committee, each participating VAMC, and the University of Maryland and of Illinois Institutional Review Boards. All studies were performed in accordance with the Declaration of Helsinki.
Table 1:
Study Cohorts and Age at Exam
| Source | Race | Gender | MS Type | N | Age at Exam |
|---|---|---|---|---|---|
| VALOMS | AA | Male | RR | 13 | 44.6 ± 2.4 |
| SP | 7 | 43.7 ± 4.1 a | |||
| Female | RR | 8 | 41.1 ± 3.0 | ||
| SP | 5 | 48.0 ± 4.5 | |||
| White | Male | RR | 24 | 48.7 ± 2.1 | |
| SP | 21 | 52.5 ± 1.5 b | |||
| Female | RR | 6 | 42.8 ± 2.8 c | ||
| SP | 7 | 52.6 ± 1.6 | |||
| ACP | AA | Male | RR | 17 | 40.4 ± 2.7 |
| Female | RR | 92 | 42.3 ± 1.1 | ||
| UCSF | White | Male | RR | 209 | 46.0 ± 0.8 |
| Female | RR | 445 | 47.0 ± 2.2 | ||
| ACP | AA | Male | Control | 11 | 49.9 ± 4.1 |
| Female | Control | 31 | 46.9 ± 3.1 | ||
| UCSF | AA | Male | Control | 100 | 45.3 ± 1.1 |
| Female | Control | 298 | 43.5 ± 0.6 | ||
| ACP & UCSF | AA | Male | Control | 111 | 45.8 ± 1.1 |
| Female | Control | 329 | 43.8 ± 0.6 | ||
| UCSF | White | Male | Control | 236 | 44.5 ± 0.9 |
| Female | Control | 425 | 42.6 ± 0.6 |
P<0.05 versus VA White male SPMS
P<0.05 versus UCSF White male RRMS
P<0.05 versus VA White female SPMS. Data is mean ± se. AA, African American; ACP, Accelerated Cure Project; RR, relapsing remitting; SP, secondary progressive.
2.2. Genomic DNA extraction from plasma samples
gDNA was extracted from plasma samples using Maxwell 16 LEV Blood DNA Kit (Promega, Madison, WI, #AS1290). Concentrations were measured using Qubit 4 Fluorometer (ThermoFisher Scientific, Waltham, MA) or Quantus Fluorometer (Promega) calibrated with double stranded DNA (ThermoFisher Scientific). Samples with concentrations below 0.3 ng/μl, or that were cloudy were amplified and repaired using the whole genome amplification REPLI-g FFPE Kit (Qiagen, Germantown, MD).
2.3. DNA analysis
Genotyping was performed using a TaqMan assay targeting the STK11 rs9282860 C/T allele (Life Technologies, Carlsbad, CA, #C_25599132_10). PCRs were performed on an Applied Biosystems ViiA7 instrument (Waltham, MA) using TaqMan Genotyping Master Mix. Data analysis was performed using ViiA7 software and Genotyper Software (Life Technologies). The genotype of a subset of samples was confirmed by PCR amplification and Sanger sequencing.
2.4. Data analysis
Group means were compared using parametric independent sample T-tests for equal or non-equal variance based on results of Levene’s test for variance. Odds ratios (ORs) were compared using Fisher’s exact test and Bonferroni correction. All analyses were carried out using SPSS v27. MS Severity Scores (MSSS) were calculated from most recent Expanded Disability Status Scale scores (EDSS) and disease duration since symptom onset as described (Roxburgh et al., 2005).
3. Results
3.1. Demographics and Clinical Features of the VALOMS patients with MS
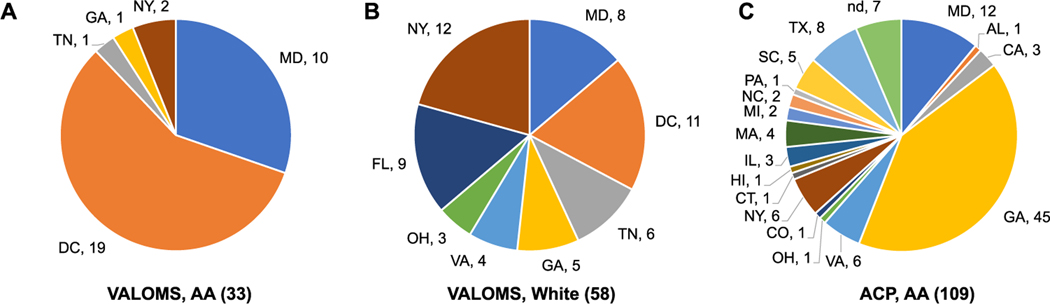
gDNA samples were obtained from PwMS from 10 different VAMCs. The majority of AA PwMS (n=33, 36%) were from the Baltimore, MD and Washington, DC areas (Fig. 1A) while the regional distribution of White PwMS (n=58, 64%) was more diverse (Fig. 1B). The total cohort had 65 males and 26 females (2.5-fold ratio); in the AA cohort the gender ratio (20M/13F, ratio = 1.5) was lower than in the White cohort (45M/13F, ratio=3.5). The age at initial exam (enrollment and blood draw) tended to be younger in AA than White PwMS (Table 1) and was significantly lower in AA compared to White male SPMS patients (43.7±4.1 vs 52.5±1.5). VALOMS SPMS patients tended to have older age at initial exam than RRMS patients, and was significantly different between White SPMS and RRMS female patients (52.6±1.8 vs 42.8±2.8).
Fig. 1.
Regional distribution of VALOMS and ACP patients with MS
Values for the number of male and female patients are shown by state for VALOMS (A) AA and (B) White PwMS; and (C) AA PwMS from ACP. nd, no data.
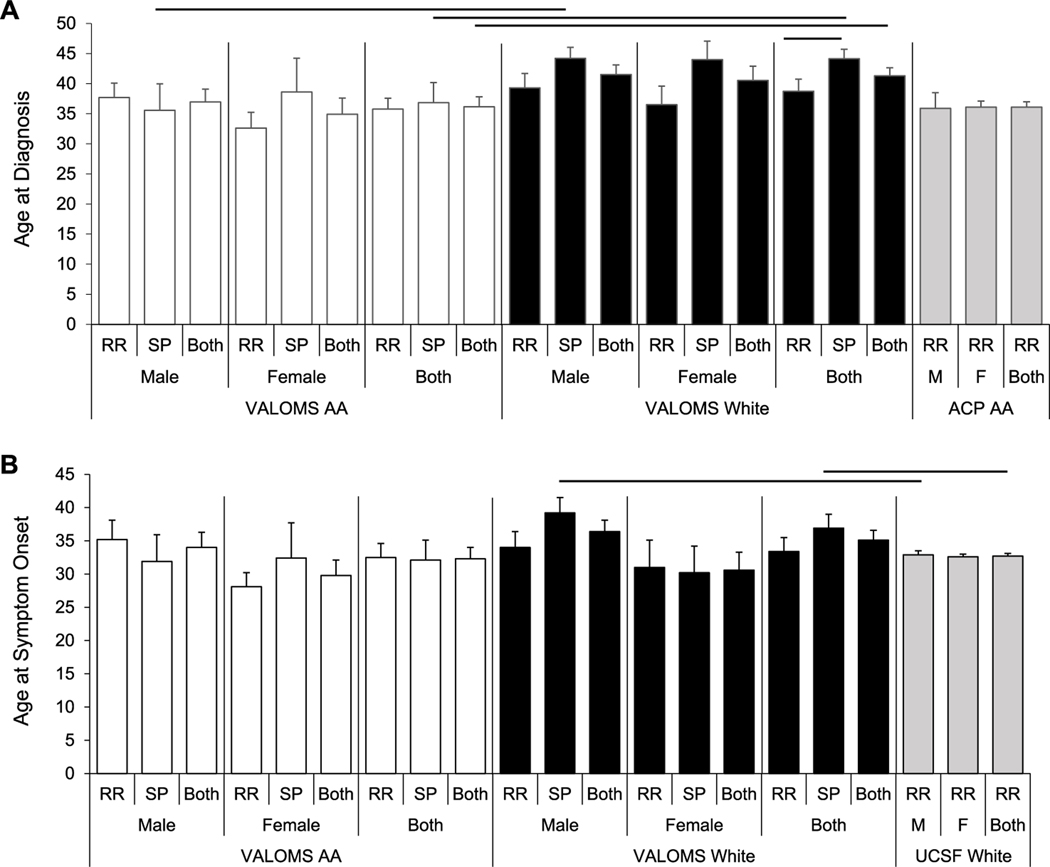
The age at diagnosis (Fig. 2A, Table S1) was younger in all AA compared to all White PwMS (36.2±1.7 vs 41.2±1.3) but did not differ between individual AA versus White genders. AA male SPMS, but not RRMS patients had a younger age at diagnosis than corresponding White SPMS patients (35.6±4.4 vs 44.2±1.9), as did the combined male and female cohorts (36.8±3.3 vs 44.1±1.6). White SPMS patients tended to have an older age at diagnosis than White RRMS patients and was significantly different between combined male and female groups (44.1±1.6 vs 38.7±2.0). The age at first symptom onset was similar between all VALOMS AA and White PwMS (Fig. 2B).
Fig. 2.
Age at diagnosis of VALOMS AA and White patients with MS
Average age at (A) diagnosis and (B) symptom onset for VALOMS AA (white bars), VALOMS White (black bars), ACP AA (gray bars in A), and UCSF White (gray bars in B) PwMS. Black lines indicate significance between groups (P<0.05). Data are mean ± se and are taken from Supplemental Table 1.
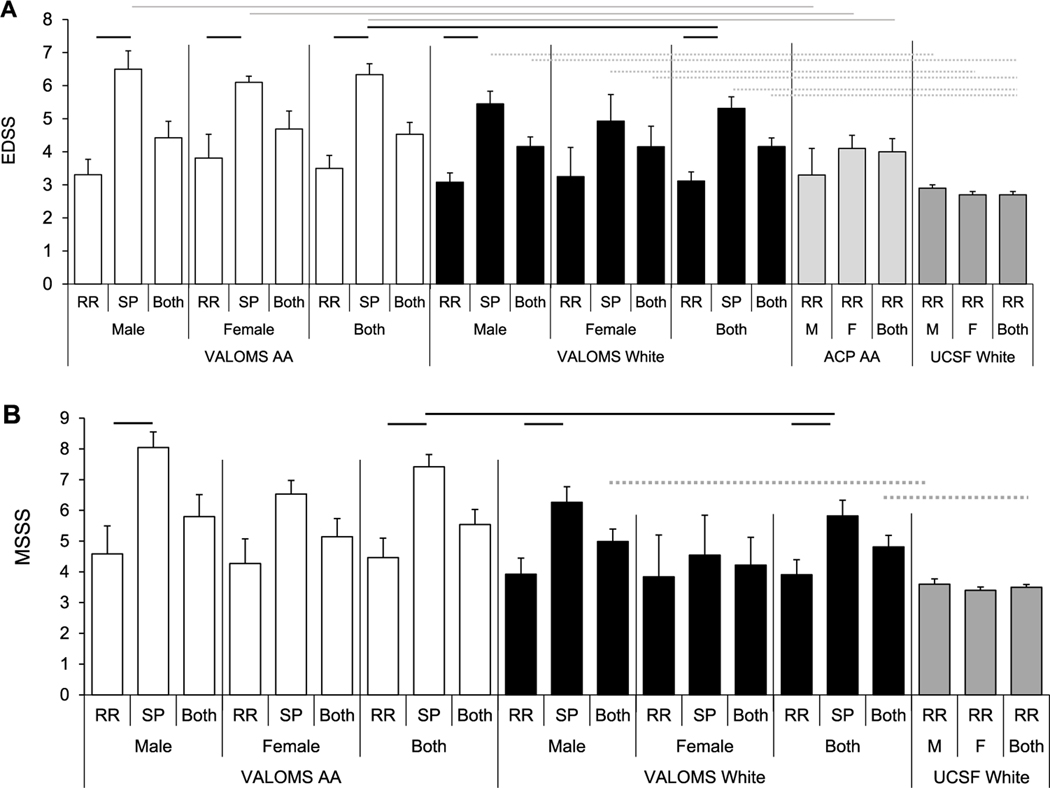
In both White and AA groups, EDSS scores were significantly higher in SPMS than corresponding RRMS patients (Table S1, Fig. 3A), except for the female White cohort. AA SPMS patients tended to have higher EDSS scores than White SPMS patients, being significantly different for combined male and female patients (6.3±0.3 vs 5.3±0.3). MSSS was higher in SPMS compared to RRMS patients (Fig. 3B), and AA SPMS patients tended to have higher MSSS than White SPMS patients, significantly different between the combined male and female cohorts (7.4±0.4 versus 5.8±0.5).
Fig. 3.
MS disease severity in AA and White patients with MS
Average (A) EDSS scores measured at last exam and (B) MSSS for VALOMS AA (white bars), VALOMS White (black bars), ACP AA (light gray bars in A), and UCSF White (dark gray bars in A and B) PwMS. Significance (P<0.05) is indicated by black lines comparing VALOMS AA and White patients; gray lines comparing VALOMS to ACP AA PwMS; and dashed lines comparing VALOMS to UCSF White PwMS. Data is mean ± se and are taken from Supplemental Table 1.
3.2. Comparison of VALOMS patients with MS to other cohorts
Comparisons were made to determine if MS diagnosis, treatment or progression differed between PwMS seen at VAMCs compared to non-VAMC facilities. The geographic distribution of AA PwMS differed between the ACP and VALOMS cohorts, with most ACP subjects from Georgia (Fig. 1C). In contrast to the VALOMS cohort, the ACP cohort consisted only of RRMS patients and was mostly females (92F/17M, ratio=5.4). Although these were RRMS patients, comparisons were made to VALOMS SPMS patients since SPMS begins as a RR disease. There were no significant differences between the average age at exam (Table 1), age at diagnosis, disease duration from diagnosis, or EDSS scores (Table S1; Fig. 3A) between the 2 groups; although as expected, VALOMS SPMS patients had higher EDSS scores than ACP RRMS patients.
Comparisons of the VALOMS White PwMS were made to the previously described UCSF cohort of 654 RRMS White PwMS (Boullerne et al., 2015), recruited from 44 different US states (Cree et al., 2009). The age at exam was similar between the groups (Table 1), although slightly higher in the VALOMS SPMS than UCSF RRMS male patients (52.5±1.5 vs 46.0±0.8). VALOMS White SPMS patients tended to have older age of symptom onset than UCSF RRMS patients with significant differences between male patients (39.2±2.3 versus 32.9±0.6) and between the combined male and female groups (36.9±2.1 versus 32.7±0.4) (Table S1, Fig. 2B). The VALOMS White SPMS female patients also had significantly longer disease duration than female UCSF White RRMS patients. VALOMS White SPMS patients had higher EDSS scores (Fig. 3A) and showed a trend toward higher MSSS (Fig. 3B) however, only the difference between the combined male and female cohorts was significant.
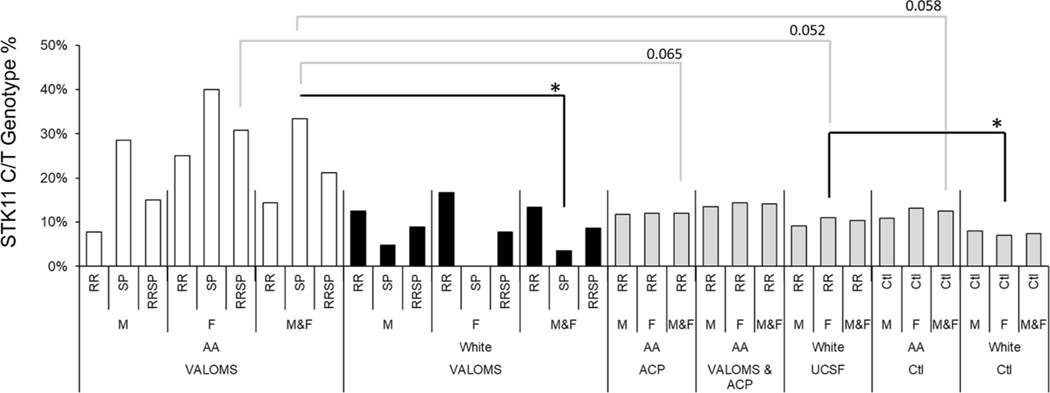
3.3. Comparison of STK11 SNP prevalence in AA patients with MS to controls
STK11 SNP prevalence did not differ between ACP and UCSF AA controls, so these were combined to yield 440 total controls. Comparison to VALOMS AA PwMS showed no significant difference in STK11 SNP prevalence (Table 2, Fig. 4), although there was a trend toward greater prevalence in female PwMS (4/13, 31% genotype frequency) versus female controls (43/329, 13%; OR=2.9, p=0.088). When stratified by subtype, the difference between all AA SPMS patients (4/12, 33%) and controls (55/440, 13%) was almost significant (OR=3.5, p=0.058). STK11 SNP prevalence tended to be greater in VALOMS compared to ACP AA PwMS, however only the difference between all VALOMS SPMS and all ACP RRMS patients was close to significant (OR=3.7, p=0.065). There were no differences between the combined VALOMS and ACP AA RRMS cohorts (n=142 total) to AA controls.
Table 2:
Prevalence of STK11 SNP in case and control groups
| STK11 Allele | Genotype % | Odds Ratio vs: | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cohort | Race | Gender | MS Type | C/T | C/C | VA White MS | ACP MS | AA Ctl | UCSF White MS | USCF White Ctl | |
| VALOMS | AA | Male | RR | 1 | 12 | 8% | 0.6 | 0.6 | 0.7 | 0.8 | |
| SP | 2 | 5 | 29% | 8.0 | 3.0 | 3.3 | 4.0 | ||||
| Both | 3 | 17 | 15% | 1.8 | 1.3 | 1.5 | 1.8 | ||||
| Female | RR | 2 | 6 | 25% | 1.7 | 2.5 | 2.2 | 2.7 | |||
| SP | 2 | 3 | 40% | n/a | 4.9 | 4.4 | 5.4 | ||||
| Both | 4 | 9 | 31% | 5.3 | 3.3 | 2.9 | 3.6 c | ||||
| Both | RR | 3 | 18 | 14% | 1.1 | 1.2 | 1.2 | 1.4 | |||
| SP | 4 | 8 | 33% | 13.5 a | 3.7 e | 3.5 b | 4.3 | ||||
| Both | 7 | 26 | 21% | 2.9 | 2.0 | 1.9 | 2.3 | ||||
| White | Male | RR | 3 | 21 | 13% | 1.4 | 1.4 | ||||
| SP | 1 | 20 | 5% | 0.5 | 0.5 | ||||||
| Both | 4 | 41 | 9% | 1.0 | 1.0 | ||||||
| Female | RR | 1 | 5 | 17% | 1.6 | 2.1 | |||||
| SP | 0 | 7 | 0% | 1.0 | 0.0 | ||||||
| Both | 1 | 12 | 8% | 1.5 | 1.0 | ||||||
| Both | RR | 4 | 26 | 13% | 1.3 | 1.7 | |||||
| SP | 1 | 27 | 4% | a | 0.3 | 0.4 | |||||
| Both | 5 | 53 | 9% | 1.2 | 1.1 | ||||||
| ACP | AA | Male | RR | 2 | 15 | 12% | 1.1 | ||||
| Female | RR | 11 | 81 | 12% | 0.9 | ||||||
| Both | RR | 13 | 96 | 12% | e | 0.9 | |||||
| VALOMS & ACP | AA | Male | RR | 5 | 32 | 14% | 1.3 | ||||
| Female | RR | 15 | 90 | 14% | 1.1 | ||||||
| Both | RR | 20 | 122 | 14% | 1.1 | ||||||
| ACP & USCF | AA | Male | Ctl | 12 | 99 | 11% | |||||
| Female | Ctl | 43 | 286 | 13% | |||||||
| Both | Ctl | 55 | 385 | 13% | b | ||||||
| UCSF | White | Male | RR | 19 | 190 | 9% | 1.1 | ||||
| Female | RR | 49 | 396 | 11% | c | 1.6 d | |||||
| Both | RR | 68 | 586 | 10% | 1.5 | ||||||
| White | Male | Ctl | 19 | 217 | 8% | ||||||
| Female | Ctl | 30 | 395 | 7% | d | ||||||
| Both | Ctl | 49 | 612 | 7% | |||||||
p=0.022
p=0.058
p= 0.052
p=0.045
p=0.065.
Fisher’s exact test. Letters are duplicated to facilitate comparisons. ACP, Accelerated Cure Project; RR, relapsing remitting; SP, secondary progressive
Fig. 4.
STK11 SNP prevalence
The C/T genotype % is shown for all groups. Significance (*, P<0.05) is indicated by black lines; near significance is indicated by gray lines. Data are taken from Table 2.
3.4. Comparison of STK11 SNP prevalence in AA to White patients with MS
Although STK11 SNP prevalence in VALOMS AA PwMS was greater than in VALOMS White PwMS (Table 2, Fig. 4), those differences did not reach significance except for prevalence in all AA SPMS patients (4/12, 33%) compared to all White SPMS patients (1/28, 4%; OR=13.5, p=0.022). Although prevalence in female AA SPMS (2/5, 40%) was greater than female white SPMS (0/7, 0%), low group sizes precluded reaching statistical significance (Fisher’s exact test = 0.15). Comparison to the UCSF White PwMS showed higher prevalence in all AA (7/33, 21%) compared to all White PwMS (68/664,10.4%), however those differences did not reach significance. The higher prevalence of the SNP in female AA PwMS (4/13, 31%) compared to female White PwMS (49/445, 11.0%) was also close to being significant (OR=3.6, p=0.052).
3.5. Comparison of STK11 SNP prevalence in VALOMS White PwMS to controls
Although SNP prevalence was slightly higher in VALOMS White RRMS patients compared to White controls (Table 2, Fig. 4), no significant differences were observed. This contrasts to UCSF White RRMS patients who had significantly higher SNP prevalence versus White female controls (Boullerne et al., 2015).
3.6. The STK11 SNP is associated with age at first symptom onset and at diagnosis
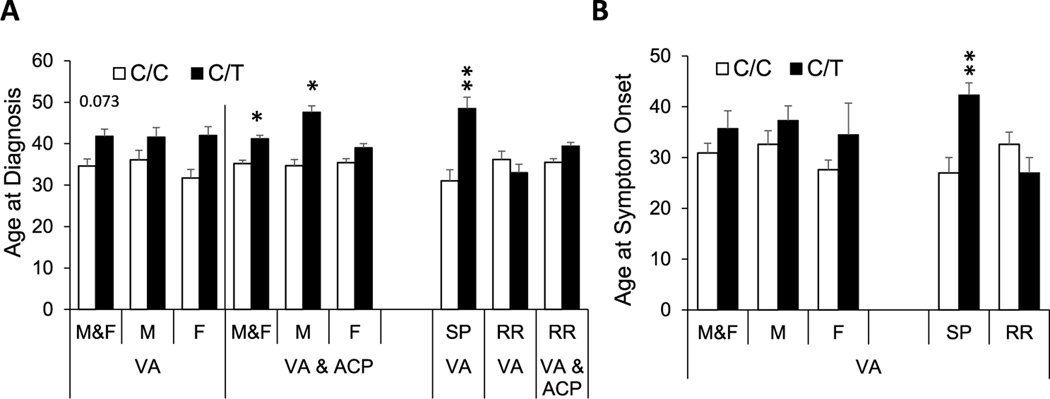
In VALOMS AA PwMS, neither average EDSS, age at first symptom onset, disease duration, nor MSSS (Table 3, Fig. 5) were different between subjects with or without the STK11 SNP; however, age at diagnosis in the combined male and female cohort tended to be older in subjects with the SNP, (41.9 ± 4.1 years) versus those without (34.6 ± 1.7 years, p=0.073). In the combined cohort of VALOMS and ACP AA PwMS, average age at diagnosis was older in those with the SNP versus those without (41.2 ± 2.5 versus 35.2 ± 0.8, p<0.05), and this association was also present in males with the SNP (47.6 ± 4.9 versus 34.7 ± 1.5, p<0.01) but not females. When stratified by MS subtype, age at first symptom onset and at diagnosis were both significantly older in SPMS subjects with the SNP, however there were no significant associations of these parameters with the SNP in RRMS subjects.
Table 3.
Effect of the STK11 SNP on clinical parameters in African American PwMS
| VALOMS AA STK11 SNP allele |
VALOMS & ACP STK11 SNP allele |
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Group | Variable | n | C/C | n | C/T | n | C/C | n | C/T |
| M&F | EDSS | 26 | 4.3 ± 0.4 | 7 | 5.2 ± 0.8 | 66 | 4.1 ± 0.3 | 9 | 5.1 ± 0.7 |
| AgeSx | 26 | 30.9 ± 1.9 | 7 | 35.7 ± 3.5 | * | ||||
| AgeDx | 26 | 34.6 ± 1.7 | 7 | 41.9 ± 4.1 a | 122 | 35.2 ± 0.8 | 20 | 41.2 ± 2.5 b | |
| MSSS | 26 | 5.3 ± 0.6 | 7 | 6.0 ± 0.8 | * | ||||
| M | EDSS | 17 | 4.2 ± 0.5 | 3 | 5.8 ± 1.5 | 22 | 4.0 ± 0.4 | 3 | 5.8 ± 1.5 |
| AgeSx | 17 | 32.6 ± 2.7 | 3 | 37.3 ± 2.9 | * | ||||
| AgeDx | 17 | 36.1 ± 2.3 | 3 | 41.6 ± 5.9 | 32 | 34.7 ± 1.5 | 5 | 47.6 ± 4.9 b | |
| MSSS | 17 | 5.4 ± 0.8 | 3 | 7.0 ± 1.5 | * | ||||
| F | EDSS | 9 | 4.7 ± 0.6 | 4 | 4.8 ± 1.0 | 44 | 4.2 ± 0.3 | 6 | 4.7 ± 0.8 |
| AgeSx | 9 | 27.6 ± 1.9 | 4 | 34.5 ± 6.2 | * | ||||
| AgeDx | 9 | 31.7 ± 2.1 | 4 | 42.0 ± 6.5 | 90 | 35.4 ± 1.0 | 15 | 39.0 ± 2.7 | |
| MSSS | 9 | 5.1 ± 0.8 | 4 | 5.3 ± 1.0 | * | ||||
| SPMS | EDSS | 8 | 6.3 ± 0.4 | 4 | 6.5 ± 0.7 | * | |||
| AgeSx | 8 | 27.0 ± 3.0 | 4 | 42.3 ± 2.4 c | * | ||||
| AgeDx | 8 | 31.0 ± 2.7 | 4 | 48.5 ± 4.3 c | * | ||||
| MSSS | 8 | 7.4 ± 0.5 | 4 | 7.5 ± 0.9 | * | ||||
| RRMS | EDSS | 18 | 3.5 ± 0.4 | 3 | 3.5 ± 1.3 | 58 | 3.8 ± 0.3 | 5 | 3.9 ± 1.0 |
| AgeSx | 18 | 32.6 ± 2.4 | 3 | 27.0 ± 3.0 | * | ||||
| AgeDx | 18 | 36.2 ± 2.0 | 3 | 33.0 ± 4.0 | 114 | 35.5 ± 0.9 | 16 | 39.4 ± 2.8 | |
| MSSS | 18 | 4.4 ± 0.7 | 3 | 4.1 ± 0.2 | * | ||||
P=0.073
P<0.05
P<0.01.
no AgeSx, MSSS, or SPMS in ACP group. Data is mean ± se. ACP, Accelerated Cure Project, EDSS, expanded disability severity scale; AgeSx, age at symptom onset; AgeDx, age at diagnosis; MSSS, multiple sclerosis severity score; RR, relapsing remitting; SP, secondary progressive.
Fig. 5.
Effect of STK11 SNP on age at diagnosis and age at symptom onset
Average (A) age at diagnosis and (B) age at symptom onset for VALOMS AA PwMS and for the combined VALOMS & ACP AA PwMS (VA & ACP), having the C/C (white bars) or C/T (black bars) allele. *, P<0.05; **, P<0.005 versus C/C. Data is mean ± se and are taken from Table 3.
In contrast to AA PwMS, White PwMS in the VALOMS cohort with the STK11 SNP (Table 4) tended to have a younger, rather than older age at first symptom onset and at diagnosis, and lower EDSS and MSSS; however, none of those differences reached statistical significance. In contrast, in the UCSF White RRMS subjects (Boullerne et al., 2015), those with the SNP had significantly lower EDSS, and male subjects showed a trend towards younger age at first symptom onset.
Table 4.
Effect of the STK11 SNP on clinical parameters in White PwMS
| VALOMS White STK11 SNP allele |
UCSF White STK11 SNP allele |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Group | Variable | n | C/C | n | C/T | n | C/C | n | C/T |
| M&F | EDSS | 52 | 4.2 ± 0.3 | 5 | 3.9 ± 1.3 | 577 | 2.8 ± 0.1 | 65 | 2.3 ± 0.2 a |
| AgeSx | 52 | 35.6 ± 1.6 | 5 | 33.6 ± 3.4 | 581 | 32.8 ± 0.4 | 66 | 32.1 ± 1.1 | |
| AgeDx | 52 | 42.1 ± 1.4 | 5 | 37.4 ± 2.5 | n/a | n/a | |||
| MSSS | 52 | 4.9 ± 0.4 | 5 | 4.3 ± 0.4 | 577 | 3.5 ± 0.1 | 66 | 3.0 ± 0.3 | |
| M | EDSS | 40 | 4.3 ± 0.3 | 4 | 3.0 ± 1.2 | 189 | 3.0 ± 0.1 | 19 | 2.1 ± 0.3 a |
| AgeSx | 40 | 37.0 ± 1.9 | 4 | 35.0 ± 4.0 | 190 | 33.4 ± 0.7 | 19 | 29.3 ± 2.4 b | |
| AgeDx | 40 | 42.2 ± 1.7 | 4 | 39.5 ± 2.6 | n/a | n/a | |||
| MSSS | 40 | 5.2 ± 0.4 | 4 | 3.0 ± 1.2 | 189 | 3.6 ± 0.2 | 19 | 3.2 ± 0.6 | |
| F | EDSS | 12 | 3.9 ± 0.6 | 1 | 7.5 | 388 | 2.7 ± 0.1 | 46 | 2.4 ± 0.2 |
| AgeSx | 12 | 30.8 ± 3.0 | 1 | 28 | 391 | 32.5 ± 0.5 | 47 | 33.2 ± 1.2 | |
| AgeDx | 12 | 41.5 ± 2.3 | 1 | 29 | n/a | n/a | |||
| MSSS | 12 | 3.8 ± 0.8 | 1 | 9.7 | 388 | 3.5 ± 0.1 | 47 | 2.9 ± 0.3 | |
| SPMS | EDSS | 26 | 5.3 ± 0.4 | 1 | 6.5 | ||||
| AgeSx | 26 | 37.2 ± 2.1 | 1 | 28 | |||||
| AgeDx | 26 | 44.5 ± 1.6 | 1 | 35 | |||||
| MSSS | 26 | 5.8 ± 0.5 | 1 | 6.3 | |||||
| RRMS | EDSS | 26 | 3.1 ± 0.3 | 4 | 3.3 ± 1.4 | ||||
| AgeSx | 26 | 33.9 ± 2.4 | 4 | 35.0 ± 4.0 | |||||
| AgeDx | 26 | 39.7 ± 2.2 | 4 | 38.0 ± 3.7 | |||||
| MSSS | 26 | 3.9 ± 0.5 | 4 | 3.9 ± 2.0 | |||||
P<0.05
P=0.088.
EDSS, expanded disability severity scale; AgeSx, age at symptom onset; AgeDx, age at diagnosis; MSSS, multiple sclerosis severity score; RR, relapsing remitting; SP, secondary progressive.
4. Discussion
In the current study we examined the prevalence of the STK11 SNP in AA and White PwMS enrolled in the VALOMS study, as well as corresponding cohorts recruited from non-VAMC facilities. Comparison of clinical features showed relatively little differences between the VALOMS patients and the other cohorts, although as expected, SPMS patients had higher neurological scores. The STK11 SNP was present at highest prevalence in AA SPMS patients, and close to be significantly greater than that in AA controls, suggesting it may be a risk factor for SPMS in this population. Interestingly, AA SPMS patients with the SNP had an older age at first symptom onset and diagnosis than those with the common allele, suggesting an influence of this SNP at an early (e.g. onset) stage of MS disease.
4.1. Military Veterans and MS
A study of Veterans with MS who had active duty during the Gulf War Era reported that MS incidence and severity is higher in Veterans compared to civilians, with the highest incidence in AAs (Wallin et al., 2012). That study found no effect of race or sex on the age of symptom onset (30.7±7.6 y) for the entire group; and consistent with that, we found no effect of race or sex on age of first symptom onset in the VALOMS cohort. However, we found that age at diagnosis tended to be younger in AA than White PwMS, with a significant difference between SPMS cohorts. We also found MSSS tended to be higher in AA than White PwMS, however only the difference between SPMS patients reached significance. This contrasts from another study which found that AA male Veterans had higher MSSS than other sex-race groups (Wallin et al., 2018). However, that finding may be due to inclusion of PPMS patients in the AA group (9.1% of AA patients) compared to only 5.1% PPMS in the White cohort; in contrast PPMS cases were not included in the current study. Overall, our results do not indicate any significant differences in disease onset or severity in AA compared to White Veterans, other than that expected for SPMS cohorts.
Comparison of the AA VALOMS to ACP AA RRMS patients showed no significant differences in either age of exam, age at diagnosis, or EDSS scores. Similarly, there were no significant differences between VALOMS and UCSF White RRMS cohorts. This suggests that Veteran PwMS at VAMCs are seen, diagnosed, and have similar disease progression as patients treated at non-VA facilities.
4.2. STK11 and MS severity and progression
Although both EDSS scores and MSSS tended to be higher in AA subjects who harbored the STK11 SNP compared to those with the C/C allele (Table 3), none of those differences reached statistical significance. This contrasts from lower MSSS and EDSS scores in the VALOMS White PwMS, and from a significant association of the SNP with lower MSSS scores in UCSF White PwMS (Boullerne et al., 2015). In the VALOMS AA PwMS, the SNP was associated with older age at disease onset and at diagnosis in all cohorts and these were significantly different in SPMS patients. In contrast, there was a trend towards an association of the SNP with younger age at onset in the UCSF White RRMS patients, which we also observed in the VALOMS White cohorts (Table 4). While small group sizes may account for lack of significant effects in the VALOMS cohorts, the trend towards an opposite association with MSSS, EDSS, disease onset, and disease diagnosis suggests that the variant differentially influences disease progression in AA compared to White PwMS.
Despite the small group size in the AA SPMS cohort (n=12 total), the STK11 SNP was almost significantly associated with MS risk compared to controls; and significantly associated when compared to all White SPMS patients. This SNP may therefore represent a risk factor for developing SPMS in the AA population. Conversely, the absence of the SNP in female white SPMS patients suggests the SNP reduces conversion to SPMS in this group; although the low group sizes precludes drawing any strong conclusions. Relatively few studies have attempted to identify risk factors for either MS progression or conversion from one form to another (Jokubaitis and Butzkueven, 2016). Environmental and dietary factors may contribute to conversion, including deficiencies in vitamin D (Ascherio et al., 2014) and rare variants in cytochrome CYP27B1 which converts Vitamin D to its active form (Ascherio, 2013, Munger and Ascherio, 2011). Smoking also has a significant association with overall MS risk as well as SPMS risk (Degelman and Herman, 2017).
4.3. STK11 SNP and disease progression
Several genetic studies have attempted to identify SNPs associated with disease progression or conversion. In contrast to the strong association of HLA with MS risk, several studies were unable to find a strong influence of HLA on MS type, progression, relapse rate, or severity (Longbrake and Hafler, 2016), although women with high HLA burden developed MS at a younger age (Isobe et al., 2016). In contrast, a retrospective study of White PwMS revealed an association of the HLA-A*02:01 allele with decreased conversion to SPMS (Misicka et al., 2020). Outside the HLA, 2 non-HLA SNPs were identified which predicted conversion from the first demyelinating event to MS and of relapse; as well as 3 non-HLA SNPs which predicted conversion and 3 which predicted relapse (Pan et al., 2016). In a comparison of ‘benign’ MS (EDSS <= 3 after 15 or more years) to ‘aggressive’ MS (EDSS >= 6 within 5 years of onset) 2 SNPs were found associated with disease course, one in CPXM2 (carboxypeptidase X); and one in IGSF9B (immunoglobulin superfamily member) (Gil-Varea et al., 2018). Variants in genes which influence mitochondrial function have also been implicated in conversion to MS or disease progression. In one study, specific mitochondrial haplotypes were found associated with increased MS risk (Tranah et al., 2015), with many SNPs present in genes encoding mitochondrial complex I. A search for variants that could distinguish different MS forms (Jia et al., 2018) showed enrichment for variants related to HSP (hereditary spastic paraplegia, a disease of upper motor neurons involving metabolic disturbance (Blackstone, 2018)) in SPMS but not RRMS patients, including 2 (REEP1 and SPG7) involved in mitochondrial function and axonal injury (Zheng et al., 2018, Atorino et al., 2003). In our EAE studies (Kalinin et al., 2020) we showed that conditional depletion of LKB1 from astrocytes, which can provide energy substrates to neurons and oligodendrocytes, reduced mitochondrial complex expression, and reduced expression of mRNAs related to astrocyte metabolism. This suggests that in PwMS, the STK11 SNP could decrease mitochondrial function or glial metabolism, either of which could contribute to increased risk of SPMS.
We found that the STK11 SNP was associated with older age at symptom onset in AA SPMS patients (Table 3). An association of older age at onset and being male with SPMS was reported in 2 studies (Misicka et al., 2020, Fambiatos et al., 2020), as well as increased risk of SPMS with older age at symptom onset, higher EDSS and faster disability accrual (Fambiatos et al., 2020). In contrast, the STK11 SNP showed a trend to younger age at onset in AA RRMS patients; as well as in VALOMS and UCSF White RRMS patients. Since SPMS begins as an RR disease, the age at first symptom onset in SPMS patients reflects the age RRMS began. A possible explanation to reconcile being a risk factor for MS yet associated with delayed onset (at least in AA patients) is that the SNP may suppress early development of MS, for example by increasing apoptosis of Tcell populations that contribute to disease onset; whereas at later times it actions in other cell types, for example reducing mitochondrial function in neurons, could accelerate RRMS to SPMS progression.
4.4. Study limitations and conclusions
The current study has several limitations. Relatively small group sizes limit the ability to detect statistically significant differences in demographic or clinical features; although an almost significant increase in STK11 SNP prevalence in AA SPMS patients versus controls was achieved with 12 SPMS patients. The geographic distribution of subjects also differs; whereas VALOMS AA PwMS were primarily from Baltimore, MD and Washington, DC areas; AA PwMS in the ACP cohort were predominantly from Southern US, while AA controls were drawn from 39 different US States (Cree et al., 2009). Since the AA genetic makeup differs across the US (Adhikari et al., 2017, Isobe et al., 2013), comparison of cohorts from different regions may be confounded by differences in African versus European ancestry.
To our knowledge, the current study represents one of the first examinations of non-HLA SNPs in a demographically diverse cohort that included military Veterans, a population with racial and ethnic diversity that reflects the US population. These findings, therefore, may be more relevant to the US population than results from larger GWAS discovery studies which primarily use Caucasian cohorts of European descent. Further study of the STK11 SNP is therefore warranted in large population-based cohorts to assess for associations with morbidity and mortality.
Supplementary Material
Acknowledgments
We thank Walter Royal, III (Morehouse School of Medicine, Georgia) and all VALOMS Investigators; David Gwynne and Sara Loud (Accelerated Cure Project), and Jorge Oksenberg and Stacy Caillier (UCSF) for assistance providing samples; Stefan Green (UIC) for assistance with Taqman assays; and Mark Maienschein-Cline (UIC) for assistance with statistical analyses.
Support
This work was supported by Merit grant I01BX002625 (DLF) and Research Career Scientist award IK6BX004852 (DLF) from the Department of Veterans Affairs; grant HC-1508-05693 from the National MS Society (MTW); and grant SI-2001-35701 from the National MS Society which supports the UCSF MS biorepository. The VALOMS study was supported by the Veterans Affairs Multiple Sclerosis Center of Excellence.
Abbreviations:
- AA
African American
- CIS
Clinically isolated syndrome
- EAE
Experimental autoimmune encephalomyelitis
- EDSS
Expanded disability severity score
- gDNA
Genomic DNA
- GWAS
Genome wide array study
- LKB1
Liver kinase B1
- MHC
Major histocompatibility complex
- MSSR
MS Surveillance Registry
- MSSS
MS severity score
- mtDNA
Mitochondrial DNA
- OR
Odds Ratio
- PPMS
Primary progressive MS
- PwMS
Patients with MS
- RRMS
Relapsing-remitting MS
- SNP
Single nucleotide polymorphism
- SPMS
Secondary progressive MS
- STK11
Serine threonine kinase 11
- VALOMS
Veterans Affairs Longitudinal MS Study
- VAMC
Veterans Affairs Medical Center
Footnotes
Declaration of Competing Interest
All authors declare that they have no competing interests.
Data availability
All data will be made available upon request after publication.
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.msard.2021.103185.
References
- International_Multiple_Sclerosis_Genetics_Consortium, 2019. Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science 365 (6460). [DOI] [PMC free article] [PubMed] [Google Scholar]
- International_Multiple_Sclerosis_Genetics_Consortium, 2018. Low-Frequency and Rare-Coding Variation Contributes to Multiple Sclerosis Risk. Cell 175 (6), 1679–1687e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranzini SE, Oksenberg JR, 2017. The Genetics of Multiple Sclerosis: From 0 to 200 in 50 Years. Trends Genet. 33 (12), 960–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patsopoulos NA, Esposito F, Reischl J, Lehr S, Bauer D, Heubach J, et al. , 2011. Genome-wide meta-analysis identifies novel multiple sclerosis susceptibility loci. Ann. Neurol 70 (6), 897–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beecham AH, Patsopoulos NA, Xifara DK, Davis MF, Kemppinen A, Cotsapas C, et al. , 2013. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat. Genet 45 (11), 1353–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isobe N, Gourraud PA, Harbo HF, Caillier SJ, Santaniello A, Khankhanian P, et al. , 2013. Genetic risk variants in African Americans with multiple sclerosis. Neurology 81 (3), 219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy JP, Cree BA, Caillier SJ, Gregersen PK, Herbert J, Khan OA, et al. , 2010. Refining the association of MHC with multiple sclerosis in African Americans. Hum. Mol. Genet 19 (15), 3080–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA, Wang J, Taylor EM, Caillier SJ, Herbert J, Khan OA, et al. , 2010. Multiple sclerosis susceptibility alleles in African Americans. Genes Immun. 11 (4), 343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cree BA, Reich DE, Khan O, De Jager PL, Nakashima I, Takahashi T, et al. , 2009. Modification of Multiple Sclerosis Phenotypes by African Ancestry at HLA. Arch. Neurol 66 (2), 226–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksenberg JR, Barcellos LF, Cree BA, Baranzini SE, Bugawan TL, Khan O, et al. , 2004. Mapping multiple sclerosis susceptibility to the HLA-DR locus in African Americans. Am. J. Hum. Genet 74 (1), 160–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beecham AH, Amezcua L, Chinea A, Manrique CP, Rubi C, Isobe N, et al. , 2020. The genetic diversity of multiple sclerosis risk among Hispanic and African American populations living in the United States. Mult. Scler 26 (11), 1329–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isobe N, Madireddy L, Khankhanian P, Matsushita T, Caillier SJ, Moré JM, et al. , 2015. An ImmunoChip study of multiple sclerosis risk in African Americans. Brain 138 (Pt 6), 1518–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amezcua L, McCauley JL, 2020. Race and ethnicity on MS presentation and disease course. Mult. Scler 26 (5), 561–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amezcua L, Rivas E, Joseph S, Zhang J, Liu L, 2018. Multiple Sclerosis Mortality by Race/Ethnicity, Age, Sex, and Time Period in the United States, 1999–2015. Neuroepidemiology 50 (1–2), 35–40. [DOI] [PubMed] [Google Scholar]
- Ventura RE, Antezana AO, Bacon T, Kister I, 2017. Hispanic Americans and African Americans with multiple sclerosis have more severe disease course than Caucasian Americans. Mult. Scler 23 (11), 1554–1557. [DOI] [PubMed] [Google Scholar]
- Cree BA, Khan O, Bourdette D, Goodin DS, Cohen JA, Marrie RA, et al. , 2004. Clinical characteristics of African Americans vs Caucasian Americans with multiple sclerosis. Neurology 63 (11), 2039–2045. [DOI] [PubMed] [Google Scholar]
- Boullerne AI, Skias D, Hartman EM, Testai FD, Kalinin S, Polak PE, et al. , 2015. A single-nucleotide polymorphism in serine-threonine kinase 11, the gene encoding liver kinase B1, is a risk factor for multiple sclerosis. ASN Neuro. 7 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan RY, Li HB, 2014. Recent progress on liver kinase B1 (LKB1): expression, regulation, downstream signaling and cancer suppressive function. Int. J. Mol. Sci 15 (9), 16698–16718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebbagh M, Olschwang S, Santoni MJ, Borg JP, 2011. The LKB1 complex-AMPK pathway: the tree that hides the forest. Fam. Cancer 10 (3), 415–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Reynolds R, Leclerc I, Rutter GA, 2011. RIP2-mediated LKB1 deletion causes axon degeneration in the spinal cord and hind-limb paralysis. Dis. Model. Mech 4 (2), 193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Luo Y, Guo W, Niu Q, Xue T, Yang F, et al. , 2017. Lkb1 maintains Treg cell lineage identity. Nat. Commun 8, 15876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh NC, Waters LR, Fowler JA, Lin M, Cunningham CR, Brooks DG, et al. , 2015. LKB1 inhibition of NF-kappaB in B cells prevents T follicular helper cell differentiation and germinal center formation. EMBO Rep. 16 (6), 753–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beirowski B, 2019. The LKB1-AMPK and mTORC1 Metabolic Signaling Networks in Schwann Cells Control Axon Integrity and Myelination: Assembling and upholding nerves by metabolic signaling in Schwann cells. Bioessays 41 (1), e1800075. [DOI] [PubMed] [Google Scholar]
- Pooya S, Liu X, Kumar VB, Anderson J, Imai F, Zhang W, et al. , 2014. The tumour suppressor LKB1 regulates myelination through mitochondrial metabolism. Nat. Commun 5, 4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen YA, Chen Y, Dao DQ, Mayoral SR, Wu L, Meijer D, et al. , 2014. Phosphorylation of LKB1/Par-4 establishes Schwann cell polarity to initiate and control myelin extent. Nat. Commun 5, 4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinin S, Meares GP, Lin SX, Pietruczyk EA, Saher G, Spieth L, et al. , 2020. Liver kinase B1 depletion from astrocytes worsens disease in a mouse model of multiple sclerosis. Glia 68 (3), 600–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallin MT, Page WF, Kurtzke JF, 2000. Epidemiology of multiple sclerosis in US veterans. VIII. Long-term survival after onset of multiple sclerosis. Brain 123 (Pt 8), 1677–1687. [DOI] [PubMed] [Google Scholar]
- Wallin MT, Page WF, Kurtzke JF, 2004. Multiple sclerosis in US veterans of the Vietnam era and later military service: race, sex, and geography. Ann. Neurol 55 (1), 65–71. [DOI] [PubMed] [Google Scholar]
- Wallin MT, Culpepper WJ, Coffman P, Pulaski S, Maloni H, Mahan CM, et al. , 2012. The Gulf War era multiple sclerosis cohort: age and incidence rates by race, sex and service. Brain 135 (Pt 6), 1778–1785. [DOI] [PubMed] [Google Scholar]
- Wallin MT, Kurtzke JF, Culpepper WJ, Coffman P, Maloni H, Haselkorn JK, et al. , 2014. Multiple sclerosis in gulf war era veterans. 2. Military deployment and risk of multiple sclerosis in the first gulf war. Neuroepidemiology 42 (4), 226–234. [DOI] [PubMed] [Google Scholar]
- Wallin MT, Culpepper WJ, Maloni H, Kurtzke JF, 2018. The Gulf War era multiple sclerosis cohort: 3. Early clinical features. Acta Neurol. Scand 137 (1), 76–84. [DOI] [PubMed] [Google Scholar]
- Wallin MT, Culpepper WJ, Campbell JD, Nelson LM, Langer-Gould A, Marrie RA, et al. , 2019. The prevalence of MS in the United States: A population-based estimate using health claims data. Neurology 92 (10) e1029–e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royal W, Wallin MT, Lee-Wilk T, Maloni H, Culpepper WJ, Finkelstein J, et al. , 2012. Clinical and Demographic Features of Participants in a Veterans Affairs Longitudinal Study of Multiple Sclerosis. Consortium of MS Centers Annual Meeting 14 (2), 75–76. [Google Scholar]
- Polman CH, Reingold SC, Edan G, Filippi M, Hartung HP, Kappos L, et al. , 2005. Diagnostic criteria for multiple sclerosis: 2005 revisions to the “McDonald Criteria”. Ann. Neurol 58 (6), 840–846. [DOI] [PubMed] [Google Scholar]
- Polman CH, Reingold SC, Banwell B, Clanet M, Cohen JA, Filippi M, et al. , 2011. Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann. Neurol 69 (2), 292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roxburgh RH, Seaman SR, Masterman T, Hensiek AE, Sawcer SJ, Vukusic S, et al. , 2005. Multiple Sclerosis Severity Score: using disability and disease duration to rate disease severity. Neurology 64 (7), 1144–1151. [DOI] [PubMed] [Google Scholar]
- Jokubaitis VG, Butzkueven H, 2016. A genetic basis for multiple sclerosis severity: Red herring or real? Mol. Cell. Probes 30 (6), 357–365. [DOI] [PubMed] [Google Scholar]
- Ascherio A, Munger KL, White R, Kochert K, Simon KC, Polman CH, et al. , 2014. Vitamin D as an Early Predictor of Multiple Sclerosis Activity and Progression. JAMA Neurol. [DOI] [PMC free article] [PubMed]
- Ascherio A, 2013. Environmental factors in multiple sclerosis. Expert. Rev. Neurother 13 (12 Suppl), 3–9. [DOI] [PubMed] [Google Scholar]
- Munger KL, Ascherio A, 2011. Prevention and treatment of MS: studying the effects of vitamin D. Mult. Scler 17 (12), 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degelman ML, Herman KM, 2017. Smoking and multiple sclerosis: A systematic review and meta-analysis using the Bradford Hill criteria for causation. Mult. Scler. Relat. Disord 17, 207–216. [DOI] [PubMed] [Google Scholar]
- Longbrake EE, Hafler DA, 2016. Linking Genotype to Clinical Phenotype in Multiple Sclerosis: In Search of the Holy Grail. JAMA Neurol. 73 (7), 777–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isobe N, Keshavan A, Gourraud PA, Zhu AH, Datta E, Schlaeger R, et al. , 2016. Association of HLA Genetic Risk Burden With Disease Phenotypes in Multiple Sclerosis. JAMA Neurol. 73 (7), 795–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misicka E, Sept C, Briggs FBS, 2020. Predicting onset of secondary-progressive multiple sclerosis using genetic and non-genetic factors. J. Neurol 267 (8), 2328–2339. [DOI] [PubMed] [Google Scholar]
- Pan G, Simpson S Jr., van der Mei I, Charlesworth JC, Lucas R, Ponsonby AL, et al. , 2016. Role of genetic susceptibility variants in predicting clinical course in multiple sclerosis: a cohort study. J. Neurol. Neurosurg. Psychiatry 87 (11), 1204–1211. [DOI] [PubMed] [Google Scholar]
- Gil-Varea E, Urcelay E, Vilariño-Güell C, Costa C, Midaglia L, Matesanz F, et al. , 2018. Exome sequencing study in patients with multiple sclerosis reveals variants associated with disease course. J Neuroinflammation 15 (1), 265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranah GJ, Santaniello A, Caillier SJ, D’Alfonso S, Martinelli Boneschi F, Hauser SL, et al. , 2015. Mitochondrial DNA sequence variation in multiple sclerosis. Neurology 85 (4), 325–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X, Madireddy L, Caillier S, Santaniello A, Esposito F, Comi G, et al. , 2018. Genome sequencing uncovers phenocopies in primary progressive multiple sclerosis. Ann. Neurol 84 (1), 51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackstone C, 2018. Hereditary spastic paraplegia. Handb. Clin. Neurol 148, 633–652. [DOI] [PubMed] [Google Scholar]
- Zheng P, Chen Q, Tian X, Qian N, Chai P, Liu B, et al. , 2018. DNA damage triggers tubular endoplasmic reticulum extension to promote apoptosis by facilitating ER-mitochondria signaling. Cell Res. 28 (8), 833–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atorino L, Silvestri L, Koppen M, Cassina L, Ballabio A, Marconi R, et al. , 2003. Loss of m-AAA protease in mitochondria causes complex I deficiency and increased sensitivity to oxidative stress in hereditary spastic paraplegia. J. Cell Biol 163 (4), 777–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fambiatos A, Jokubaitis V, Horakova D, Kubala Havrdova E, Trojano M, Prat A, et al. , 2020. Risk of secondary progressive multiple sclerosis: A longitudinal study. Mult. Scler 26 (1), 79–90. [DOI] [PubMed] [Google Scholar]
- Adhikari K, Chacón-Duque JC, Mendoza-Revilla J, Fuentes-Guajardo M, Ruiz-Linares A, 2017. The Genetic Diversity of the Americas. Annu. Rev. Genomics Hum. Genet 18, 277–296. [DOI] [PubMed] [Google Scholar]
- Isobe N, Damotte V, Lo R,V, Ban M, Pappas D, Guillot-Noel L, et al. , 2013. Genetic burden in multiple sclerosis families. Genes Immun. 14 (7), 434–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.