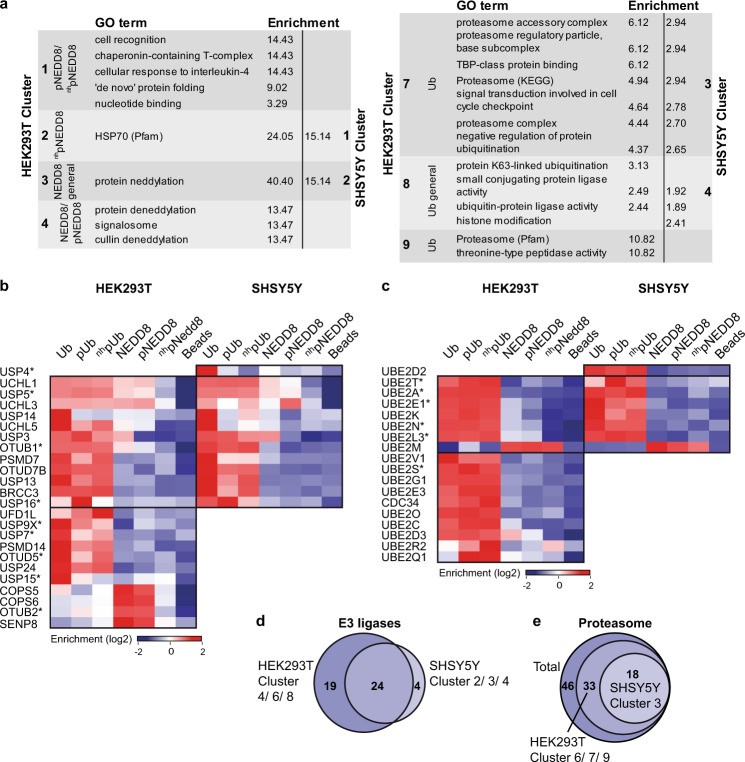
Fig. 5. Enzymes of Ub/NEDD8 conjugation/deconjugation and their binding preferences.
a GO terms, KEGG, and Pfam annotations with enrichment in the identified clusters (Fig. 4b) including biological processes, protein complexes, and families. For HEK293T clusters 5 and 6, none of the GO terms was enriched. b Hierarchical clustering of DUBs identified in HEK293T and SHSY5Y AE-MS experiments. The DUBs with an asterisk were found previously to be either activated or inhibited by phosphorylation at S6536. c Hierarchical clustering of E2 enzymes identified. The E2s with an asterisk were studied before with phosphorylated Ub as a substrate19. Thresholds for the ANOVA statistics (b,c) were set as follows: FDR = 0.02, S0 = 1. Enrichment is indicated in red, whereas lack of enrichment is indicated in blue. n = 3 biologically independent experiments measured in technical duplicates. d Venn diagram overview showing all E3s found in either one of the cell lines or in both AE-MS experiments. In which clusters (Fig. 4b) the E3s are mainly represented in is indicated. e Venn diagram showing the amount of proteasomal subunits found in the two experiments and the clusters (Fig. 4b) they are represented in. Source data are provided as a Source Data file.