Figure 2.
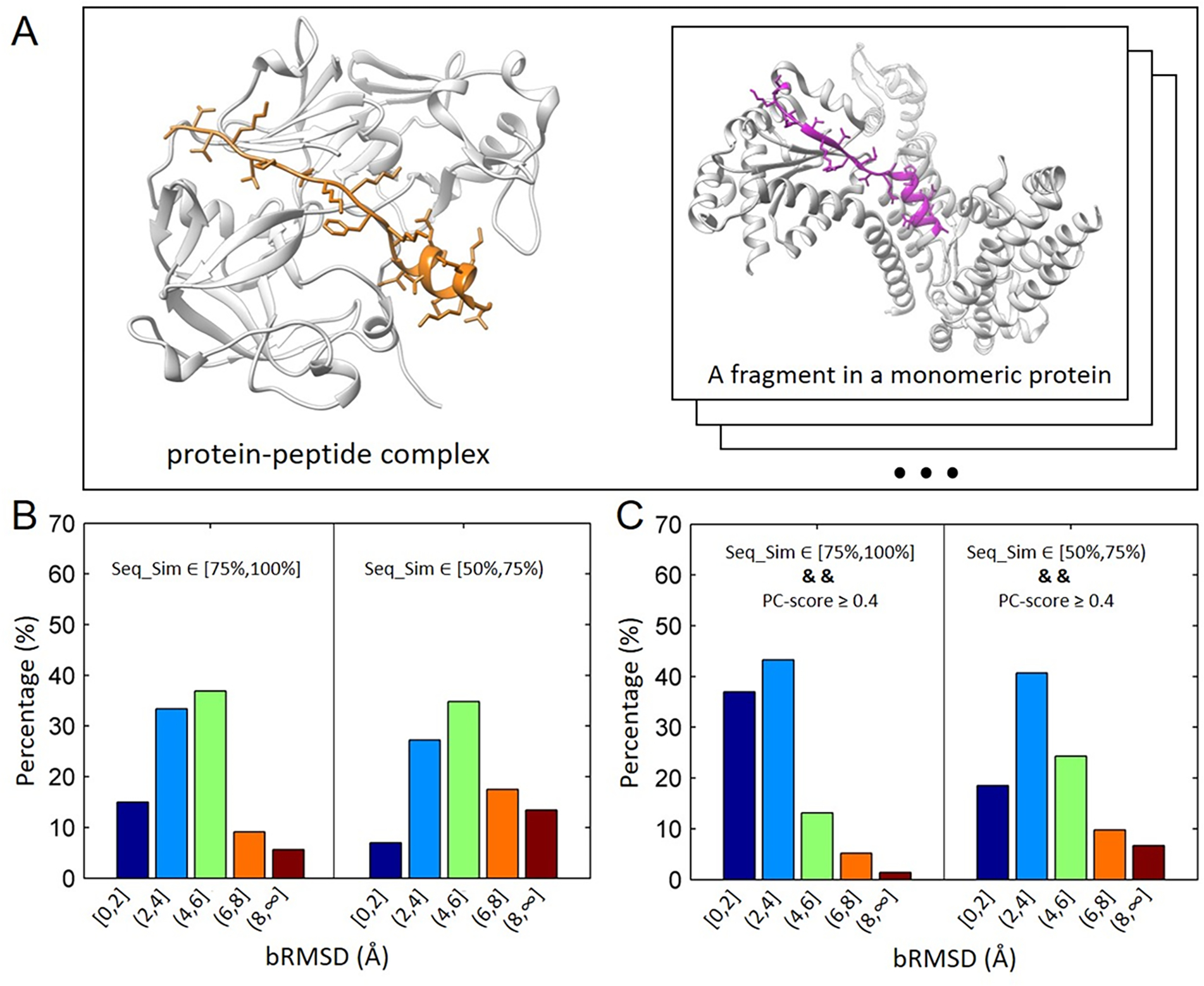
Structural comparison of peptides in protein-peptide complexes and corresponding fragments in monomeric proteins. (A) Left panel: an example of protein-peptide complex (PDB: 1avf) with the peptide sequence length 22 (color in orange); Right Panel: a fragment from a monomeric protein (PDB: 4xgc, chain C, residues 259–280). bRMSD of the peptide and the fragment hit is 2.3 Å. (B) Distributions of bRMSDs between peptide structures and corresponding fragments with two ranges of sequence similarities, [75%, 100%] and [50%, 75). The calculation was based on 89 non-redundant protein-peptide complexes in PepPro benchmark. (C) Similar to (B) except that fragments used for calculation have similar environments (or similar interacting interface, PC-score ≥ 4.0) with query peptides.
