Figure 4.
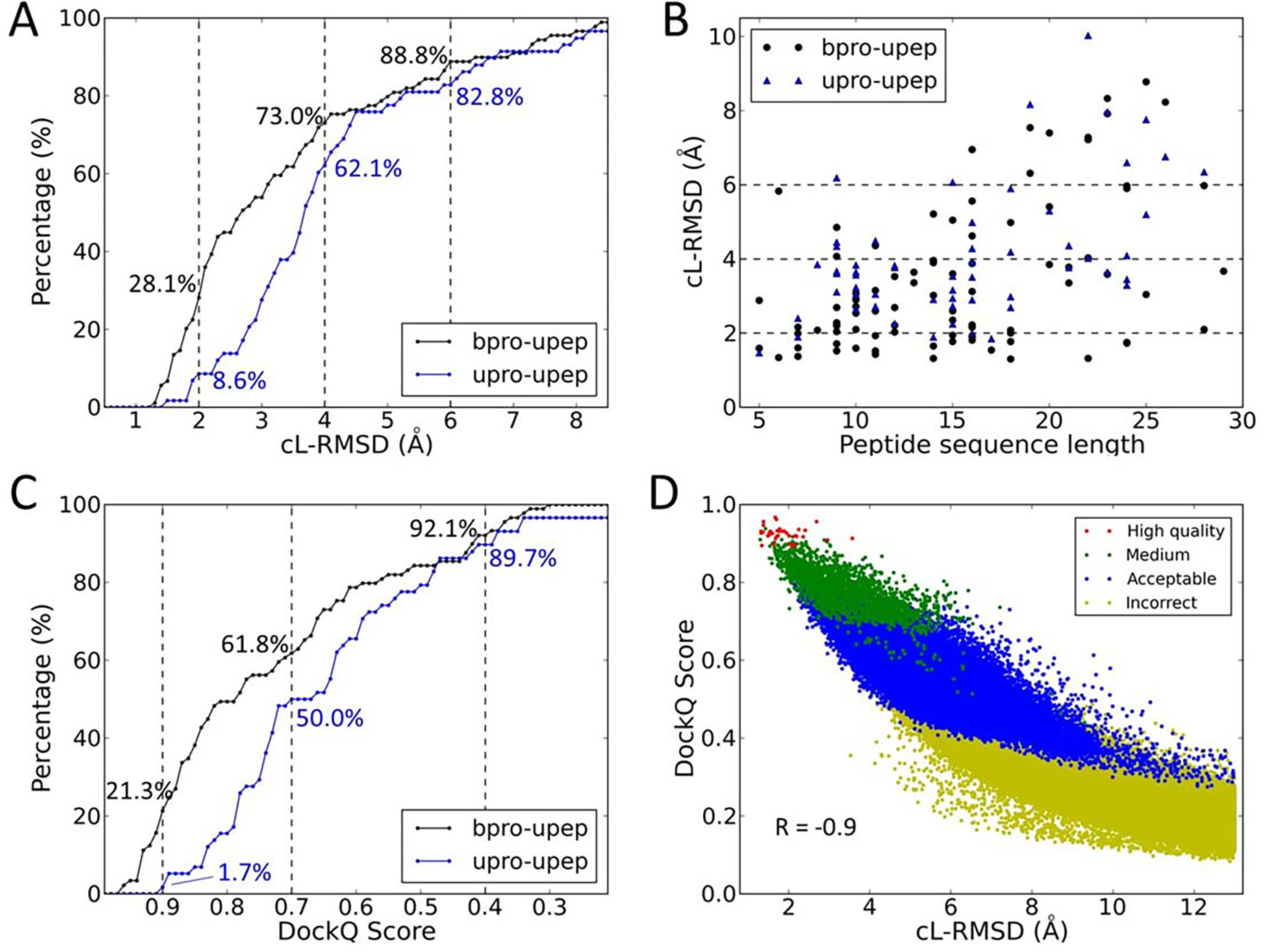
The performance of binding mode sampling for both the bound docking cases and unbound docking cases in PepPro.
(A) The success rates of peptide binding mode sampling using different values of cL-RMSD as the thresholds for the bound docking cases (bpro-upep) and unbound docking cases (upro-upep). Dashed lines represent 2.0 Å, 4.0 Å, and 6.0 Å.
(B) The lowest cL-RMSD value of sampled models for each complex for both the bound docking and the unbound docking. The peptide sequence length is shown for each complex.
(C) Sampling success rates using different values of DockQ Score as the thresholds for the bound and unbound dockings. Dashed lines represent DockQ Scores of 0.9, 0.7, and 0.4.
(D) Distributions of sampled models (bound docking) in three evaluation metrics, cL-RMSD (x-axis), DockQ Score (y-axis), and capri_metric (colors).
