Figure 6.
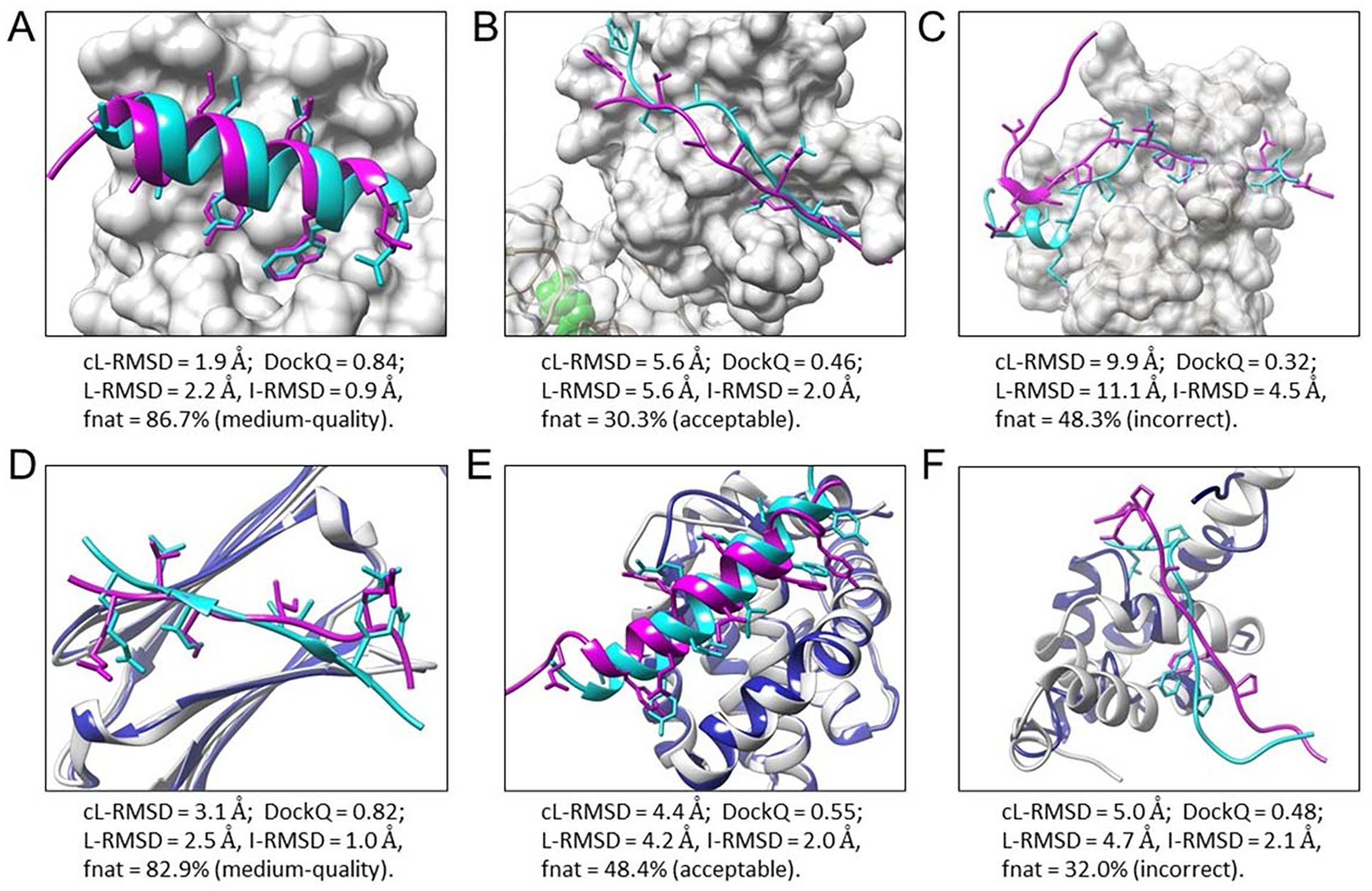
Six examples of peptide binding modes predicted by MDockPeP2. Experimental bound peptide structures are colored cyan, and predicted peptide binding modes are colored magenta. Side chains of contact peptide residues are represented by the stick model. Proteins in the bound docking cases (A-C) are displayed by the surface and colored light gray. For the unbound docking cases (D-F), unbound protein structures (colored blue) are superimposed on bound protein structures (colored light gray). (A) The top predicted model of PDB 1t0j. (B) A predicted model (no. 289) of PDB 2whx. (C) A low-quality model of PDB 4ext. (D) A predicted model (no. 8) of PDB 4m5s, using the unbound protein structure PDB 3utk. (E) A predicted model (no. 260) of PDB 3kj0, using the unbound protein structure PDB 2mhs. (F) A predicted model (no. 228) of PDB 3kut, using the unbound protein structure PDB 1g9l.
