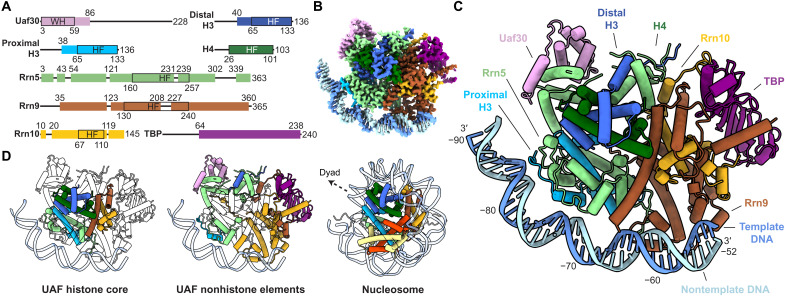
Fig. 1. Structure of UAF with TBP and promoter DNA.
(A) Domain organization of UAF subunits. Residues built are represented as colored boxes with ranges indicated on top and domain boundaries (black outlines) indicated below. WH, winged helix; HF, histone fold. Loops L1 and L2 of the Rrn5 HF, helices ɑ1 to ɑ2 of the Rrn9 HF are extended relative to histone proteins H4 and H2B, respectively. The Rrn10 HF is missing the corresponding helix ɑ3 of H2A. (B) Cryo-EM density map of UAF, postprocessed with DeepEMhancer (45). (C) Atomic model represented as pipes (ɑ helices), planks (β strands), ribbons (DNA backbone), and sticks (DNA bases). The respective subunits are labeled and colored as in (A). (D) Histone-like hexameric core [left, HF domains outlined in (A)] and non–histone-like periphery (middle) of UAF, colored by subunit. The budding yeast nucleosome [Protein Data Bank (PDB), 1ID3] (26) is shown in the same orientation on the right, with the dyad axis indicated. Corresponding histones are colored as in UAF.