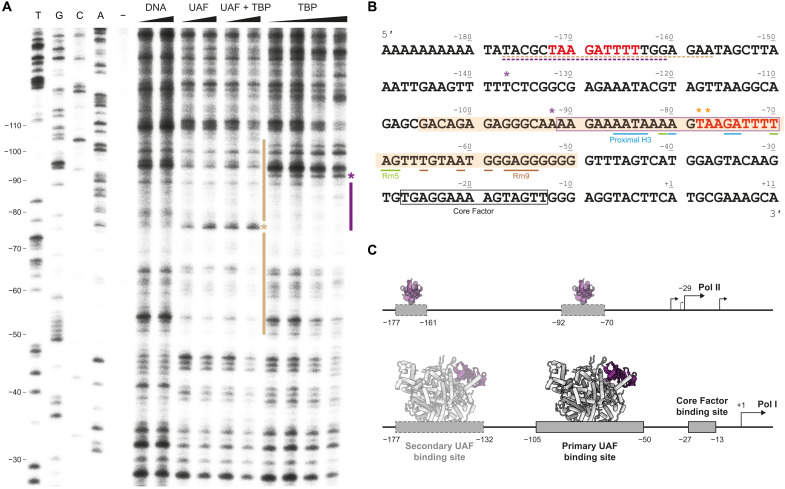
Fig. 3. In vitro DNase I footprint of UAF on promoter DNA.
(A) DNase I footprint visualized by primer extension using the nontemplate strand from position +25 of the 35S rRNA promoter. From left to right: Sanger sequencing reactions (T, G, C, and A), untreated DNA (−), free DNA treated with 0.36 and 0.72 U of DNase I (DNA), DNA bound to UAF (4 and 8 μM), UAF and TBP (4 and 8 μM, 1:1 ratio), and TBP (4, 8, 16, and 32 μM), treated with 0.72 U of DNase I. Sequencing lanes are labeled according to the nontemplate strand. Bars indicate protection. Asterisks indicate an increase in sensitivity to DNase I. (B) Annotated 35S rRNA promoter sequence. The nontemplate strand sequence is displayed. Shaded region indicates the primary UAF binding site. Boxed regions indicate a potential TBP binding site as suggested by the footprint (purple) and known Core Factor binding site (14) (black). Potential upstream UAF and TBP binding sites identified in fig. S7A are underlined. Bases contacted by UAF in the structure are indicated. Colored in red is the putative repeat sequence bound by Rrn5 and the proximal H3. (C) A schematic model of UAF and TBP binding at the 35S rRNA gene promoter. In the absence of UAF, TBP binding to DNA enables Pol II–mediated transcription from upstream of the Pol I transcription start site (2, 55). In the presence of UAF, UAF binding is favored; TBP is sequestered from DNA to support Pol I recruitment.