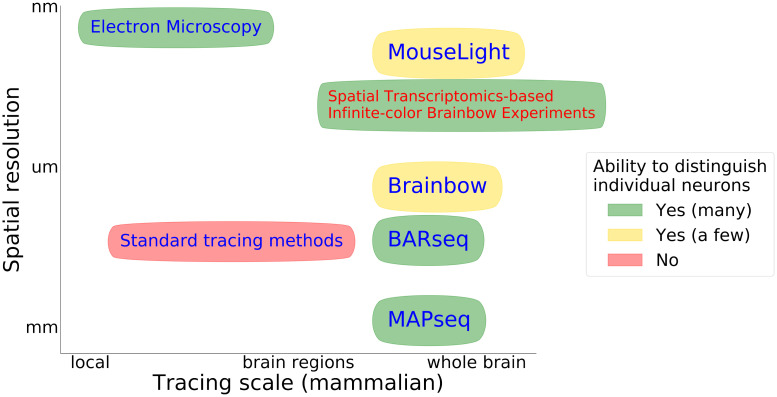
Fig 1. Some existing methods for neuronal morphology mapping.
Here we non-exhaustively summarize the landscape of existing neuronal tracing methods. The horizontal axis indicates the typical scale of these neuronal tracing methods, ranging from local tracing to mapping axonal projections across the whole brain; the vertical axis indicates optical resolution in the typical images obtained from these methods. The ellipse color represents the number of neurons that can be traced and distinguished; red indicates that the method cannot readily distinguish between spatially nearby neurons, yellow indicates that the method can be used to trace several individual neurons, and green indicates the possibility of simultaneously mapping thousands or millions of neurons. Here ‘standard tracing methods’ refer to approaches such as Golgi staining and viral tracers; cf. Section 2 for more details. Note that many of these methods rely on conventional optical microscopes, and their resolution can potentially be increased via expansion microscopy and/or super resolution microscopy, though this may incur additional experimental costs. In this paper we focus on the possibility of extending molecular barcoding methods by increasing the signal density to allow for more fine-grained morphological reconstructions; we call such experiments Spatial Transcriptomics-based Infinite-color Brainbow Experiments (STIBEs).