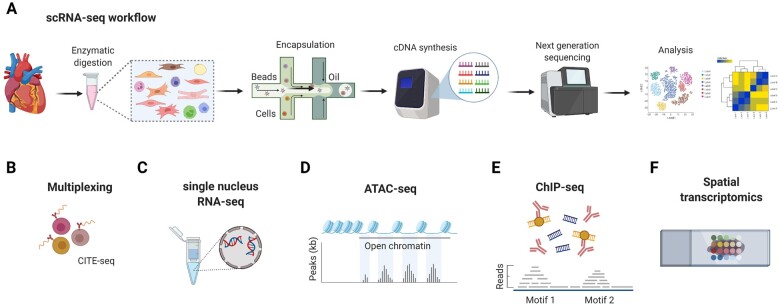
Figure 1.
Overview of scRNA-seq and related technological advancements to study the cardiac transcriptome. (A) The technologies used for scRNA-seq mostly employ microfluidics platforms for single-cell encapsulation as initial step of sample preparation, combined with next-generation sequencing approaches for entire transcriptome measurements.7 (B) To improve the precision of immune cell subset identification, which might be relevant for rare cell populations, oligonucleotide-barcoded antibody labelling, or lipid-tagged indices for barcoding of cells prior to their processing for scRNA-seq can be used.8 , 9 This multimodal measurement of cell surface labels in combination with transcript expression allows combining several biological samples in one scRNA-seq library, which is referred to as multiplexing. This reduces inter-sample technical bias and facilitates exclusion of cellular doublets, thereby limiting analytical artefacts. (C) An alternative to scRNA-seq is the single nucleus RNA-seq (snRNA-seq) method, which might be preferable when working with tissues that are difficult to dissociate or frozen and large cells such as cardiomyocytes.10 , 11 Other technical advancements involve combined methods for epigenetic profiling with scRNA-seq, such as (D) assay for transposase-accessible chromatin using sequencing (ATAC-seq) or (E) chromatin immunoprecipitation followed by sequencing (ChIP–seq).7 ATAC-seq allows sequencing of the regions of the genome with open or accessible chromatin, while ChIP-seq enables genome-wide profiling of DNA-binding proteins and histone modifications. (F) Novel developments combine transcriptomics with spatial information, in order to overcome the issue that the information on cellular distribution gets lost due to the generation of single-cell suspensions.12