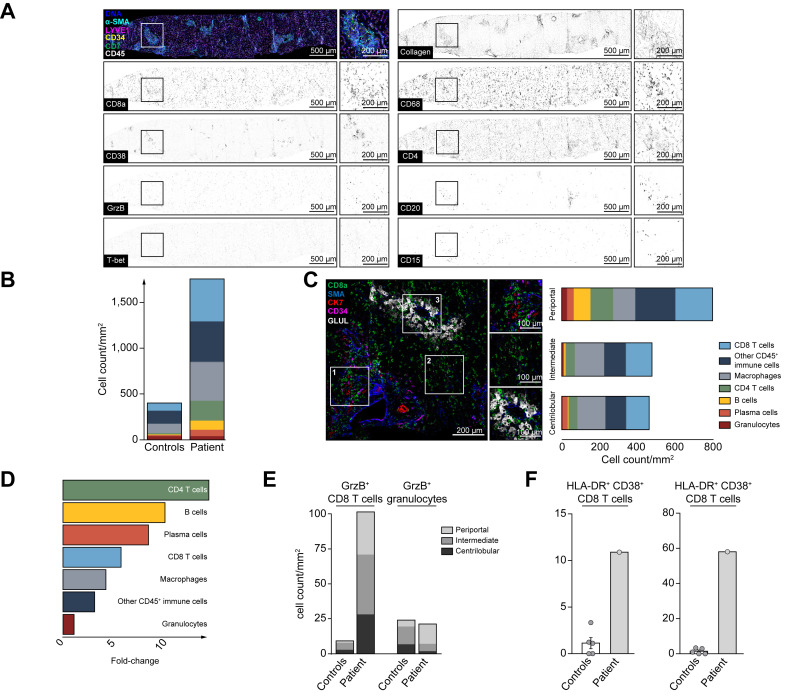
Fig. 2.
Deep spatial analysis of the hepatic immune environment reveals immune infiltration dominated by cytotoxic CD8 T cells.
The liver biopsy was analyzed by highly multiplexed IMC. (A) Pseudocolored IMC images of the liver biopsy. Top left: Composite visualization of CD45, CK7, CD34, LYVE1, a-SMA and DNA are indicated by colors. Single marker visualization is shown for markers depicted on the left border of the visualizations. A region of interest is indicated by the box and detailed at higher resolution. Scale bars indicate 500 μm and 200 μm in the inserts, respectively. (B) Cell counts of immune cell subsets in the liver are shown as stacked barplots compared to controls. (C) Composite IMC image visualizing examples of liver zones defined by structural markers: Periportal (top), intermediate (middle) and centrilobular zones (bottom). Immune cell counts within the defined regions visualized as stacked barplots next to the respective inserts. Scale bars indicate 200 μm and 100 μm in the inserts, respectively. (D) Enrichment of indicated immune populations was calculated by dividing cell density in the patient sample by the mean of control samples. (E) Distribution of GrzB-positive CD8 T-cell and granulocyte cell counts within the liver zones. (F) Frequency (left) and cell density (right) of HLA-DR+ CD38+ CD8 T cells are shown for the patient with hepatitis and controls. GrzB, granzyme B; IMC, imaging mass cytometry.