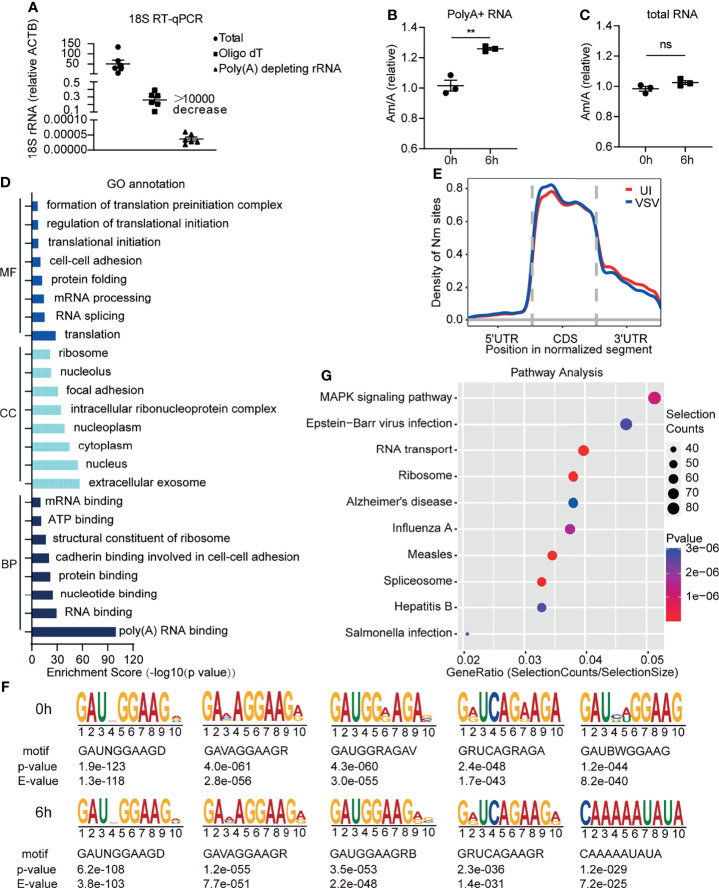
Figure 1.
Increased Am modification on poly A+ RNA in macrophages upon viral infection. (A) Determination of poly A+ RNA purity through RT-qPCR with primers specific to 18S rRNA and Actb. Total, total RNA; Oligo dT, poly A+ purified by Oligo dT beads; Poly A+ depleting rRNA, poly A+ purified by Oligo dT beads and rRNA depleting kit (n=6); (B) Quantification of the Am/A ratio in poly A+ RNA of RAW264.7 cells with or without VSV infection (n=3). 0h, RNA from RAW264.7 cells; 6h, RNA from RAW264.7 cells infected with VSV (MOI=1) for 6 h; (C) Quantification of the Am/A ratio in total RNA of RAW264.7 cells with or without VSV infection (n=3). 0h, RNA from RAW264.7 cells; 6h, RNA from RAW264.7 cells infected with VSV (MOI=1) for 6 h; (D) GO terms of MF, CC, BP for Nm-methylated transcripts of RAW264.7 at the steady state; MF, Molecular function; CC, Cell Compartment; BP, Biological process; (E) Metagene profile of Nm sites distribution along a normalized mRNA transcript; (F) Sequence logo of enriched motifs behind Nm sites identified by HOMER; (G) KEGG pathway enrichment for Nm up-regulated genes after viral infection. UI, poly A+ RNA from RAW264.7 cells; VSV, poly A+ RNA from RAW264.7 cells infected with VSV (MOI=1) for 6 h. All data are mean ± SEM of biologically independent samples.ns, not significant, **P < 0.01, two-tailed unpaired Student’s t test.