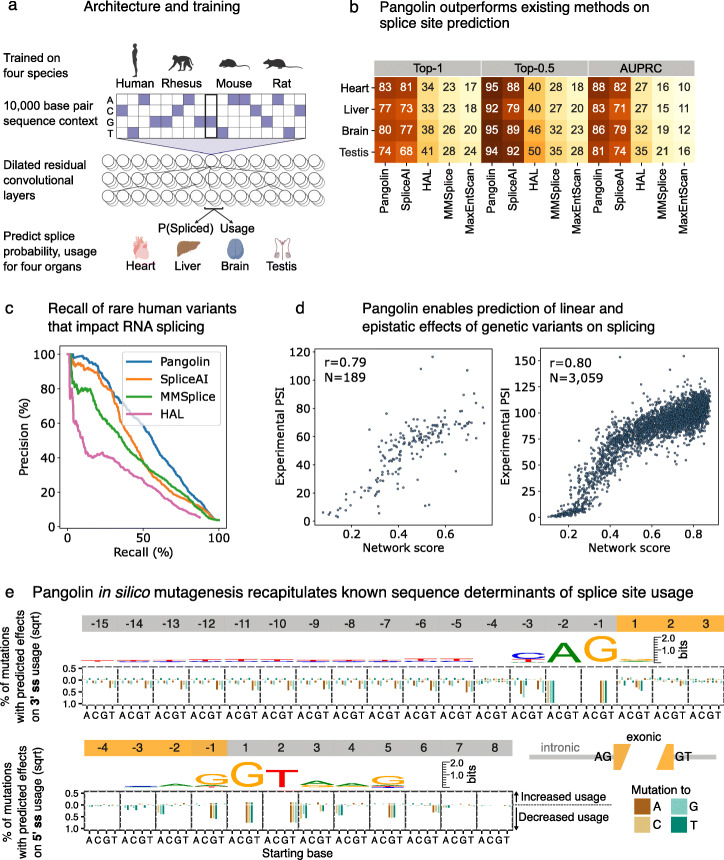
Fig. 1.
Overview of Pangolin and evaluation. a Schematic and architecture of Pangolin. b Heatmap summarizing the performance of Pangolin, SpliceAI, HAL, MMSplice, and MaxEntScan with respect to three metrics including top-1 accuracy. c Precision-recall curves representing the precision and recall from multiple methods for the prediction of splice-disrupting variants as identified in Cheung et al. [8] (1050 splice-disrupting variants out of 27,733 total). d Scatter plots showing measured versus predicted effects of single genetic variants (left) or a combination of genetic variants (right) on RNA splicing. Measured effects of single genetic variants and combinations of variants were obtained from Julien et al. [15] and Baeza-Centurion et al. [3] respectively. e In silico mutagenesis of 6416 exons from human chromosomes 7 and 8. Barplots show for each base the percent of mutations (square root) predicted to increase or decrease usage by at least 0.2