Figure 1.
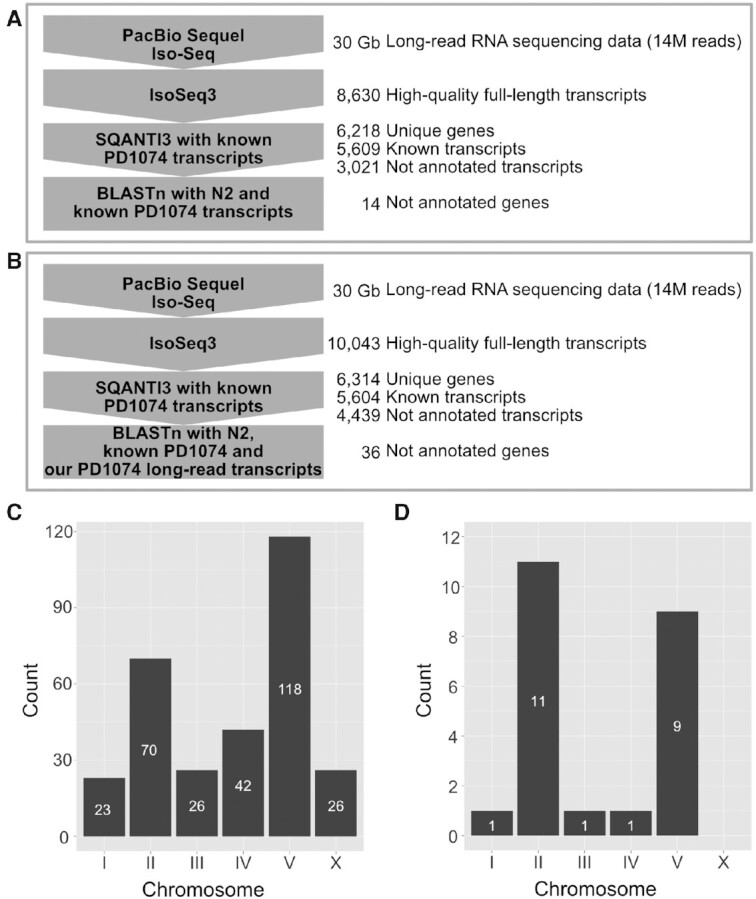
Schematic overview of data analysis to identify previously non-annotated genes and chromosomal distribution of strain-specific genes. (A and B) Computational workflows for finding previously non-annotated genes of (A) PD1074 and (B) CB4856 using long-read RNA sequencing. PacBio Iso-Seq data were processed using IsoSeq3 to produce high-quality full-length transcripts, SQANTI3 to extract newly detected gene candidates by comparing the transcripts with known PD1074 transcripts and BLASTn to verify the candidates by searching for them in either (A) the N2 and PD1074 known gene databases or (B) the N2 and PD1074 known gene databases supplemented with our long-read PD1074 transcripts database. (C and D) Chromosomal distribution of (C) PD1074- and (D) CB4856-specific genes.
