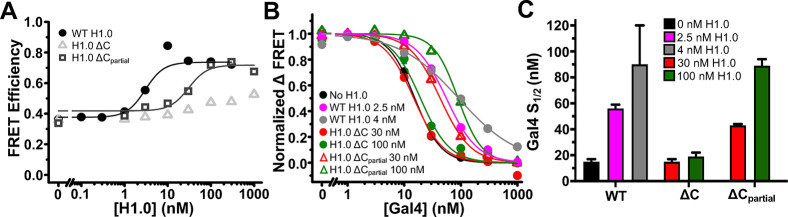
Figure 5.
FRET measurements of H1.0 CTD truncations binding to nucleosomes and regulating Gal4 binding within DNA-OctFRET nucleosomes. (A) FRET efficiency of nucleosomes with increasing amounts of H1.0. WT H1.0 increases FRET efficiency as seen before and binds with an S1/2 of 3 ± 1 nM (black circles). H1.0 ΔC increases nucleosome FRET efficiency only slightly over the range of concentrations that bind in an EMSA, indicating that the mutant has little effect on linker DNA wrapping (gray triangles). H1.0 ΔCpartial increases the FRET efficiency of nucleosomes to a similar degree as WT H1.0 with a reduced S1/2 of 30 ± 10 nM showing that it alters the nucleosomal DNA wrapping to a similar degree as WT H1.0 but binds nucleosomes with a reduced affinity (gray squares). (B) Normalized ΔFRET efficiency of nucleosomes bound to H1.0 with increasing amounts of Gal4. Gal4 binding to nucleosomes alone (black) and nucleosomes bound with two different H1.0 ΔC concentrations (red and green circle) is similar, showing H1.0 ΔC does not alter Gal4 binding. In the presence of WT H1.0 (pink and gray circles) or H1.0 ΔCpartial (red and green triangles), Gal4 binding to nucleosomes is reduced with S1/2 values reported in panel C, indicating H1.0 ΔCpartial is able to alter Gal4 binding to nucleosomes. (C) S1/2 values for Gal4 binding data in panel B. Gal4 binding to nucleosomes alone (black): 15 ± 2 nM. Gal4 binding to nucleosomes with 2.5 nM WT H1.0 (pink): 56 ± 3 nM. Gal4 binding to nucleosomes with 4 nM WT H1.0 (gray): 90 ± 30 nM. Gal4 binding to nucleosomes with 30 nM H1.0 ΔC (red): 15 ± 2 nM. Gal4 binding to nucleosomes with 100 nM H1.0 ΔC (green): 19 ± 3 nM. Gal4 binding to nucleosomes with 30 nM H1.0 ΔCpartial (red): 43 ± 1 nM. Gal4 binding to nucleosomes with 100 nM H1.0 ΔCpartial (green): 89 ± 5 nM.