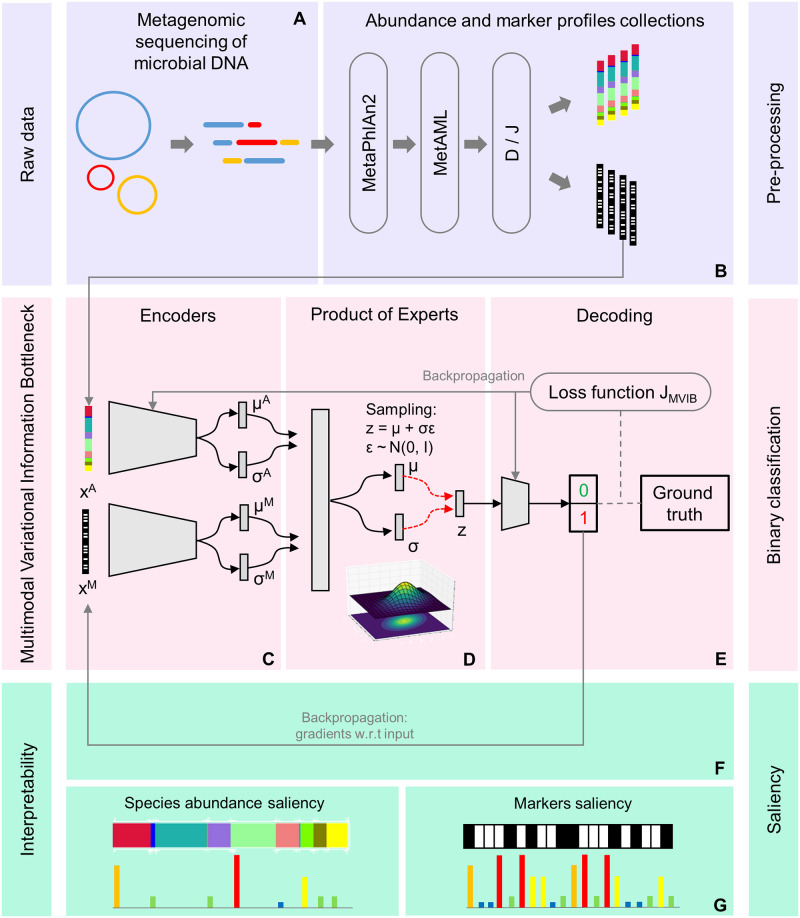
Fig 1. Full workflow.
(A) The raw data, i.e., shotgun metagenomic sequencing data of the human gut microbiome. (B) For pre-processing, we leverage MetaPhlAn2 and MetAML to extract species-relative abundances and strain-level markers. We consider two pre-processing schemes to produce two different dataset collections, default (D) and joint (J). (C) A high-level representation of the probabilistic encoders of the MVIB model. (D) The Product of Experts computes a single joint posterior i.e. z; the joint posterior z is sampled with the reparametrisation trick [28]. (E) A logistic regression decoder estimates the probability of whether a subject is affected by a certain disease. (F, G) The gradients of the output class are computed with respect to (w.r.t) the input vectors and used to compute saliency maps.