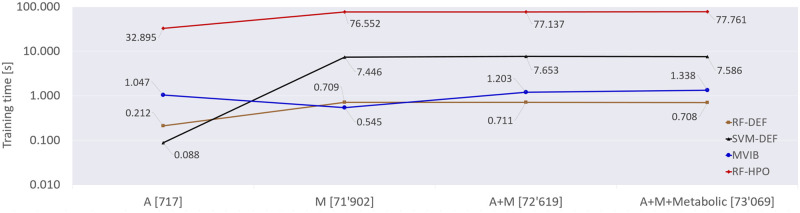
Fig 7. Training time comparison across different machine learning models and various feature space dimensions.
The values reported in this graph consist in the training time measured in seconds. The Colorectal-Metabolic dataset extracted from [37] has been used for training all models. The depicted training times are obtained by averaging the run times of five experiments with different random train/test splits. A: species-relative abundance profiles. M: strain-level marker profiles. Metabolic: metabolite profiles. RF-DEF: Random Forest with default Scikit-learn implementation. SVM-DEF: Support Vector Machine with default Scikit-learn implementation. RF-HPO: Random Forest with hyperparameter optimisation (see S1 File). For MVIB, the JMVIB objective has been adopted for the optimisation (Eq 5). On the x-axis, next to the modality name, the feature space dimension is reported in square brackets.