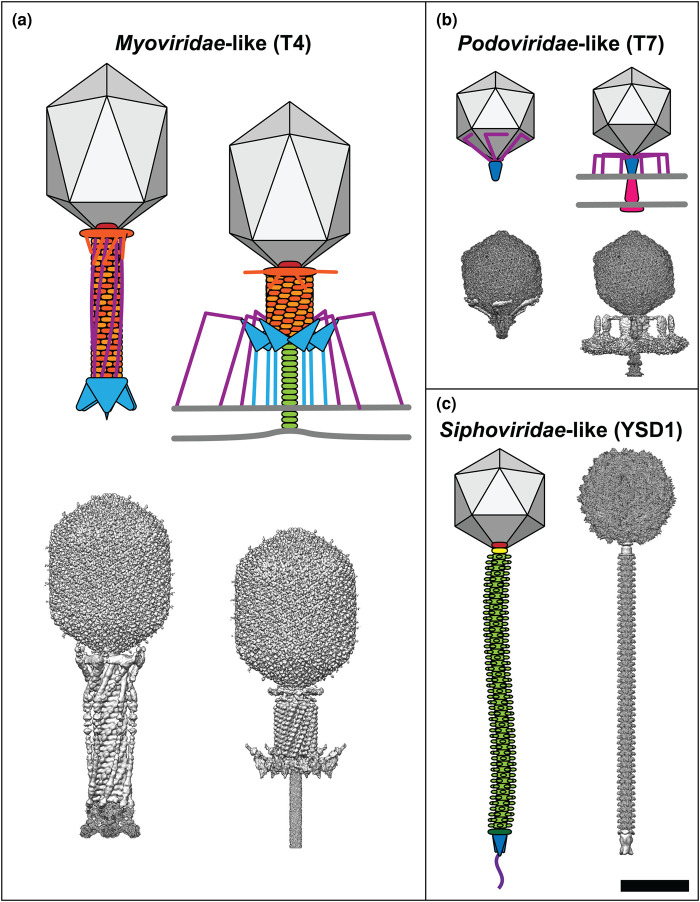
Figure 1. Comparison of tailed phage morphology.
For each family of tailed phages a cartoon schematic is shown alongside a composite of the corresponding cryo-EM reconstructions (where available). Phages are drawn to scale and their component parts are coloured according to function: capsid, grey; portal, red; tail sheath, orange; tail terminator, yellow; tail tube, light green; distal tail protein, dark green; baseplate, cyan; tail spike, blue; tail fibres, purple; ejectosome, pink. (a) Myoviridae-like phages such as T4 have contractile tails, (b) Podoviridae-like viruses such as T7 have very short non-contractile tails and assemble an ejectosome for genome delivery, and (c) Siphoviridae-like phages such as YSD1 have long, flexible non-contractile tails. T4 maps before host attachment and genome ejection: capsid, EMD-6323 [25]; tail and sheath, EMD-1126 [26]; baseplate, EMD-3374 [27]. T4 maps after host attachment and sheath contraction: capsid, EMD-6323 [25]; sheath, EMD-5528 [28]; baseplate, EMD-3396 [27]; tail tube, EMD-8786 [29]. T7 maps before host attachment and genome ejection: capsid, EMD-30139 [30]; portal–tail complex, EMD-31319 [31]; tail fibres, EMD-31315 [31]. T7 maps after host attachment and genome ejection: capsid, EMD-30139 [30]; portal–tail complex, EMD-31321 [31]; tail fibres, EMD-31318 [31]; ejectosome, EMD-22680 [32]. YSD1 maps before host attachment and genome ejection: capsid, EMD-22182 [33]; portal/connector, unpublished data; tail, EMD-22183 [33]; spike, unpublished data. Scale bar = 50 nm. Composite maps were generated in UCSF Chimera version 1.14 [34].