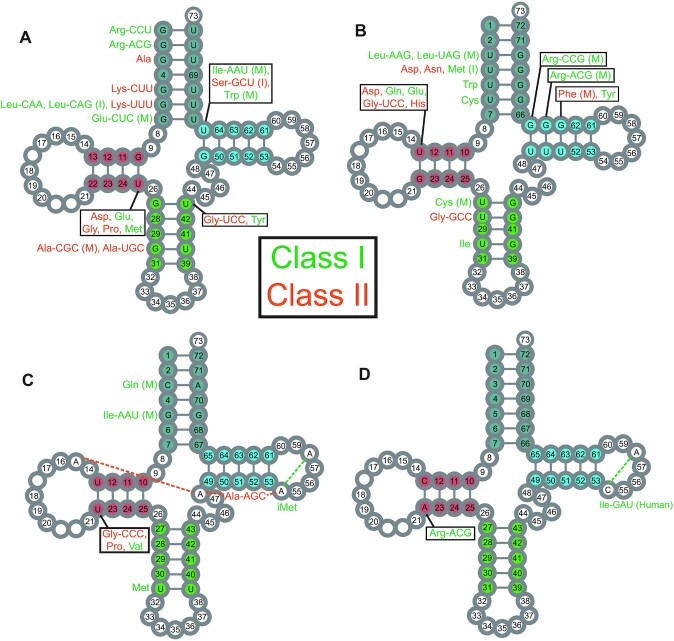
Figure 3.
Summary of GoU base pairs and other non-Watson–Crick base pairs in helical stems of tRNAs. Isotype- and position-specific conserved GoU (A) and UoG (B) base pairs in tRNA stems, as well as other non-Watson–Crick pairs (C, D) are shown by Sprinzl position. The long-arm tRNAs are characterized by a G13oA22. (C) In the AA-stem, Gln-UUG tRNAs present often in Mammalia a CoA opposition at positions 3:70 and Ile-AAU tRNAs have GoG at positions 5:68. Gly-CCC, Pro, and Val tRNAs have a UoU at positions 13:22 in the D-stem. In addition, Ala-AGC tRNAs have an RoA tertiary interaction at positions 15:48 together with A54oA58 (47), and Met tRNAs have AoA at positions 54:58. (D) Arg-ACG tRNAs have a CoA at positions 13:22 in the D-stem, and H. sapiens Ile-GAU tRNAs have the unusual CoA tertiary interaction at positions 54:58 in the T-loop. Numbers in stem positions indicate that no non-Watson–Crick interactions are observed at that position in insect or mammalian tRNAs (except for tRNA-Thr at positions 4:69 where both GoU and UoG are observed). Amino acids are colored based on aminoacyl tRNA synthetase class (Class I in green and Class II in orange). (M) indicates that the base pair is observed in Mammalia and (I) indicates that the base pair is observed in Insecta (all valid for Drosophila species). Isodecoders without either letter are conserved in both insects and mammals.