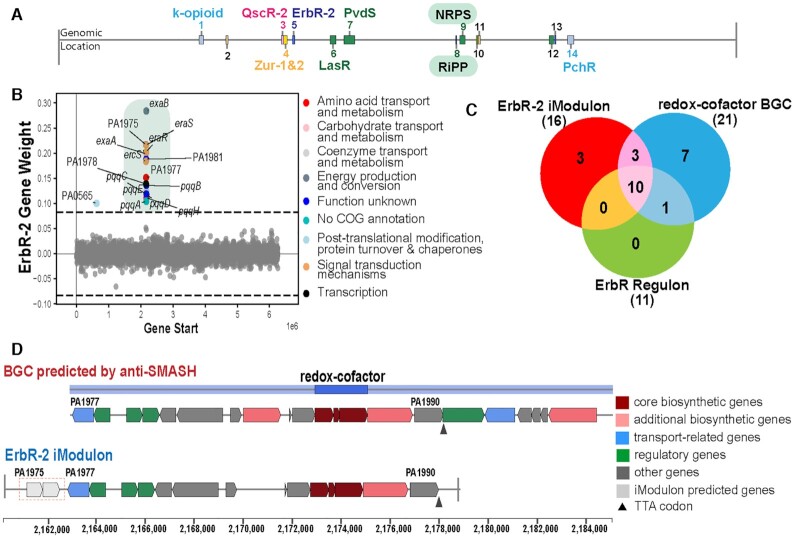
Figure 3.
iModulons can aid in the definition of genomic boundaries of biosynthetic gene clusters (BGCs). (A) Genomic locations of the 14 predicted BGCs in the P. aeruginosa PAO1 by using the anti-SMASH software. (B) Scatter Plot showing the gene weights of the ErbR-2 iModulon with the color depicting the COG categories of the genes that it contains. (C) Venn diagram depicting the status of the genes in the ErbR-2 iModulons, ErbR regulon, and the predicted redox-cofactor BGCs by using the anti-SMASH software. (D) Genomic overview of the redox-cofactor BGCs predicted by the anti-SMASH software, alongside the iModulons whose boundaries are defined by genes between the PA1975-PA1990.