Figure 5.
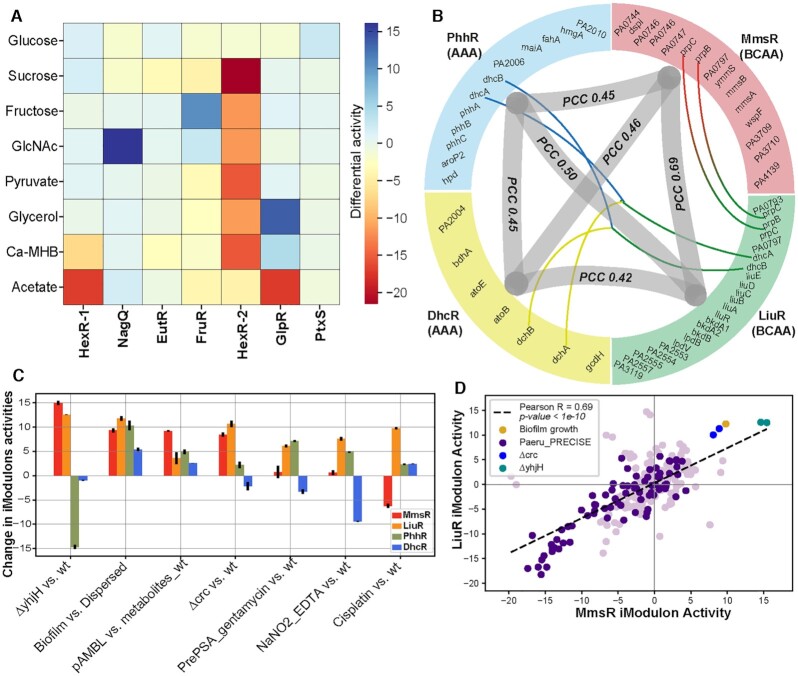
iModulons related to Carbon metabolism and Amino acid/Nucleotide metabolism. (A) Heat map depicting the differential activity of glucose, sucrose, fructose, N-acetylglucosamine, pyruvate, glycerol, Ca-MHB (bacteriological media), and acetate with respect to HexR-1, NagQ, EutR, FruR, HexR-2, GlpR, and PtxR iModulons. (B) Correlation plot among the Branched chain amino acid [BCAA (LiuR and MmsR)] and the Aromatic amino acid [AAA (DhcR and PhhR)]. The outer layer is divided into the four arcs which depict the four different iModulons. Thin lines represent the common genes among the iModulons, and the thick line connecting different iModulons depicts the Pearson correlation coefficients (PCC). (C) Bar plot representing the iModulon activities of MmsR, LiuR and PhhR under different conditions. The x-label shows some conditions used in the study. The ‘△yhjH vs. wt’ is the knockout of the yhjH, ‘Biofilm vs. Dispersed’ is the biofilm mode of growth, ‘pAMBL vs. metabolite_wt’ is the pAMBL plasmid showing overexpression of metabolites, ‘△crc vs. wt’ is the deletion of the global regulator of crc, ‘PrePSA_gentamycin vs. wt’ is the pre-PatH-Cap library of P. aeruginosa (‘PSA’ PAO1-GFP) treated with gentamycin,’NaNO2_EDTA vs. wt’ is the presence of sodium nitrite and EDTA in the media, and ‘Cisplatin vs. wt’ is the presence of cisplatin and bile in the media. (D) Scatter plot showing the correlation between the BCAA pathways iModulons, i.e. LiuR and MmsR, with the PCC of 0.69.