Figure 7.
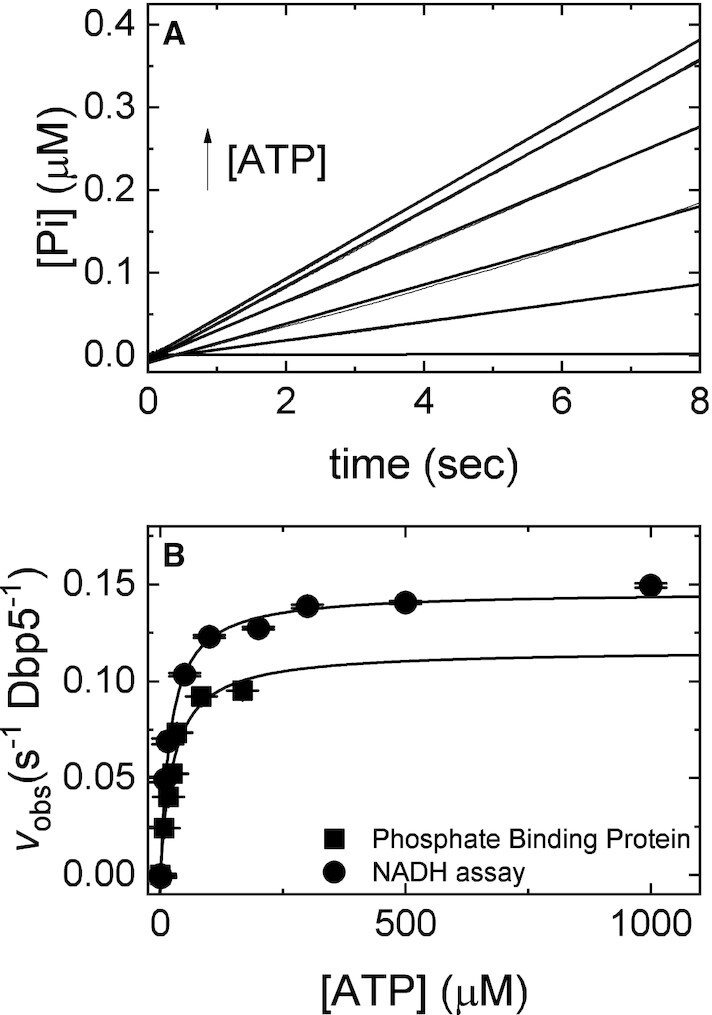
Direct measurement of Pi release from Gle1–Dbp5 via PiBP. (A) Time courses of phosphate release in a pre-equilibrated mixture of 1 μM Dbp5 (0.5 μM after mixing) and 20 μM Gle1 (10 μM after mixing) upon rapid mixing with various [ATP] (0, 5, 10, 15, 20, 50, 100 μM after mixing) containing 6 μM PiBP (3 μM after mixing). Continuous lines through the data are the best fits to a linear equation. (B) [ATP]-dependance of the observed steady-state Pi release in A (solid squares) or steady-state ATP hydrolysis in the presence of the NADH regenerating system (26,31,49). Continuous lines through the data are the best fits to a rectangular hyperbola yielding the maximum velocity per enzyme (kcat= 0.15 ± 0.02 s–1 Dbp5–1, circles; kcat= 0.12 ± 0.01 s–1 Dbp5–1, squares) from the amplitude and KM (20 ± 3 μM, circles; 26 ± 6 μM, squares) from the [ATP] at half-maximum velocity (Table 1). Uncertainty bars represent standard errors in the fits and are contained within the data points. InsP6 is included in all experiments at an equimolar concentration with Gle1.
