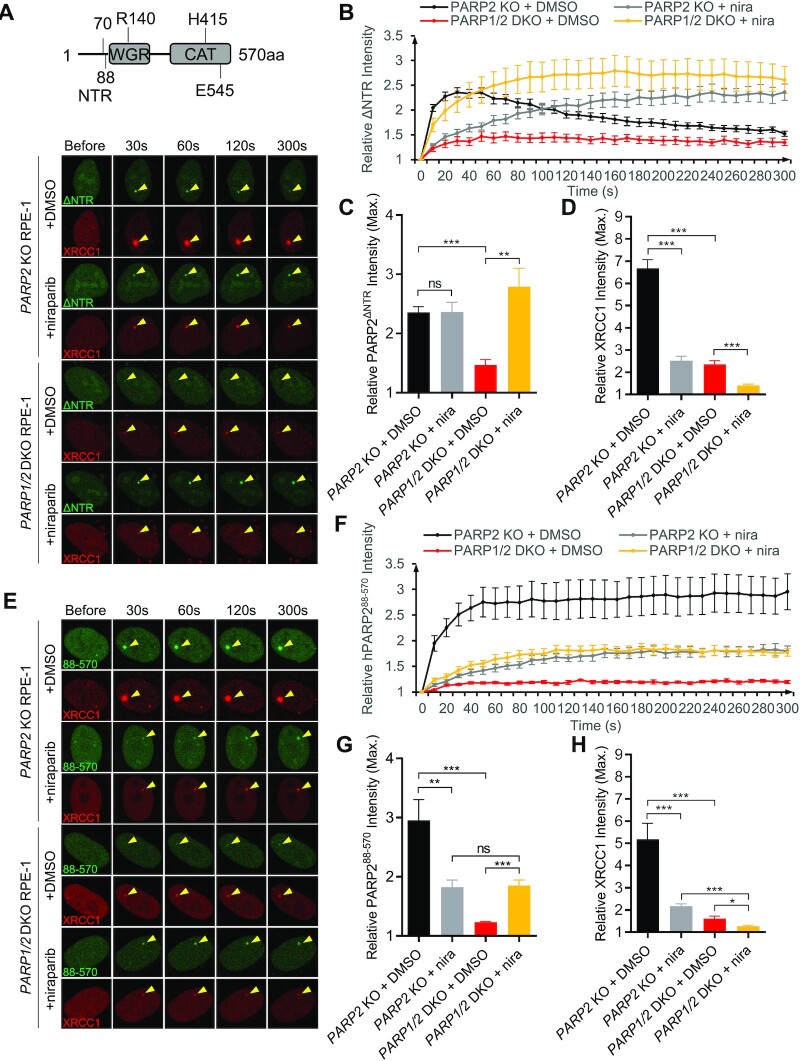
Figure 5.
The NTR is largely dispensable for PARP2 recruitment and trapping by niraparib. (A) The schematic of PARP2 domain with aa number marked for human PARP2. Representative images of GFP-SV40NLS-ΔNTR-PARP2 and mRFP-XRCC1, and (B) the relative intensity kinetics of GFP-SV40NLS-ΔNTR-PARP2 at DNA damage sites in PARP2 KO and PARP1/2 DKO RPE-1 cells in the presence and absence of niraparib. (C, D) The maximal relative intensity of (C) GFP-SV40NLS-ΔNTR-PARP2 and (E) mRFP-XRCC1. (E) Representative images of GFP-SV40NLS-PARP288-570 and mRFP-XRCC1, and (F) the relative intensity kinetics of GFP-SV40NLS- PARP288-570 at DNA damage sites in PARP2 KO and PARP1/2 DKO RPE-1 cells in the presence and absence of niraparib. (G and H) The maximal relative intensity of (G) GFP-SV40NLS- PARP288-570, and (H) mRFP-XRCC1. The dots and bars represent means and SEM, respectively, from one representative experiment out of 2–4 with n > 8 cells each time with consistent results. The two-tailed unpaired Student's t-test was used to calculate p-values. ns, P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001.