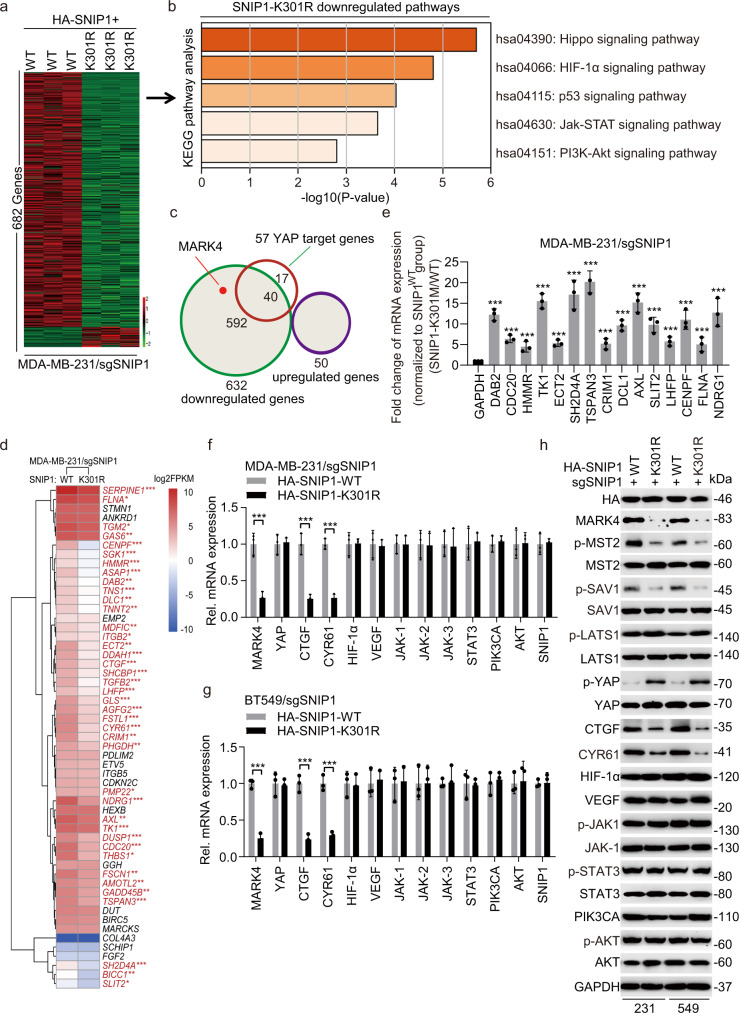
Fig. 3. KMT5A-mediated SNIP1 K301 methylation activates Hippo/YAP signaling.
a Heatmap of RNA-Seq analysis of differentially expressed genes downregulated by SNIP1-K301R (SNIP1K301R) (twofold change and FDR < 0.05). SNIP1 sgRNA-resistant Flag-SNIP1WT and SNIP1K301R mutant were re-expressed in sgSNIP1/MDA-MB-231 cells. b Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis indicates that the genes downregulated by the SNIP1K301R mutant were predominantly associated with the Hippo signaling pathway. c Venn diagram shows YAP target genes that are regulated by SNIP1 demethylation (SNIP1K301R). d Heatmap representing expression levels of 55 YAP target genes determined by RNA-Seq in (a). Forty genes were found to be significantly downregulated in K301R mutant cell lines were highlighted in red. e qPCR analysis of a group of YAP target genes that were induced upon SNIP1 K301 methylation (SNIP1K301M) (n = 3). f, g these representative typical pathway genes that may be regulated by SNIP1 methylation were measured and verified in MDA-MB-231 (f) and BT549 (g) cells by qRT-PCR (n = 3). h Western blot was performed in MDA-MB-231/sgSNIP1 and BT549/sgSNIP1 cells transfected with SNIP1 sgRNA-resistant Flag-SNIP1WT and SNIP1K301R mutant (n = 3). Data information: In (d, e–g), statistical analysis was performed by a two-tailed t-test. mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001. Panels (e–h) show 1 experiment representative of three independent experiments with similar results.