Figure 1.
IMD regulates the intestinal progenitor-cell transcriptome
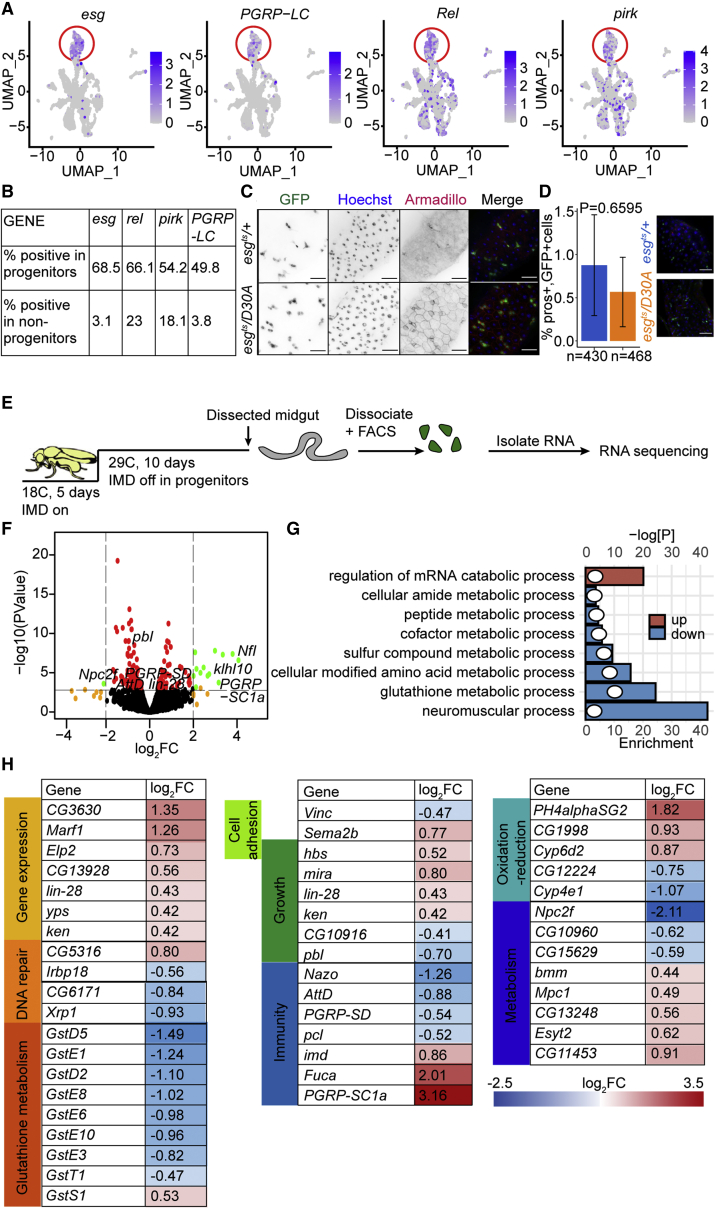
(A) Uniform manifold approximation and projection (UMAP) plot of intestinal epithelial cells isolated from 10-day-old esgts flies showing expression of the progenitor marker esg and the IMD pathway components PGRP-LC, Rel, and pirk. Progenitors are circled in red.
(B) Quantification of the percentage of progenitors and non-progenitors that express the indicated genes.
(C) Visualization of GFP, DNA (Hoechst), and the beta-catenin ortholog in intestines of 10-day-old esgts/+ and esgts/UAS-imdD30A (esgts/D30A) flies. Scale bars represent 25 μm.
(D) Quantification of the percentage of GFP-positive cells that express the enteroendocrine cell marker prospero in esgts/+ (n = 20) and esgts/D30A (n = 22) flies. esgts/+ (n = 430) and esgts/D30A (n = 468) n = number of GFP+ cells. Representative images are shown with DNA in blue, esg+ cells in green, and prospero-positive enteroendocrine cells in red. Significance measured using Student’s t test. Scale bars represent 25 μm.
(E) Schematic representation of an experimental strategy to quantify gene expression in progenitor cells purified from esgts/+ and esgts/D30A flies. Each experiment was performed in triplicate.
(F) Volcano plot showing relative changes (x axis) and significance (y axis) in gene expression of purified progenitors from esgts/D30A flies compared with age-matched esgts/+ controls. Genes are color-coded to indicate significance and relative gene expression changes.
(G) Gene Ontology analysis of processes significantly affected by inhibition of IMD in progenitors. Column size indicates the degree of enrichment for each term, and dots indicate the log-transformed significance of the respective enrichment.
(H) Representative sample of genes with affected expression upon inhibition of IMD in progenitors. See also Figures S1 and S2.