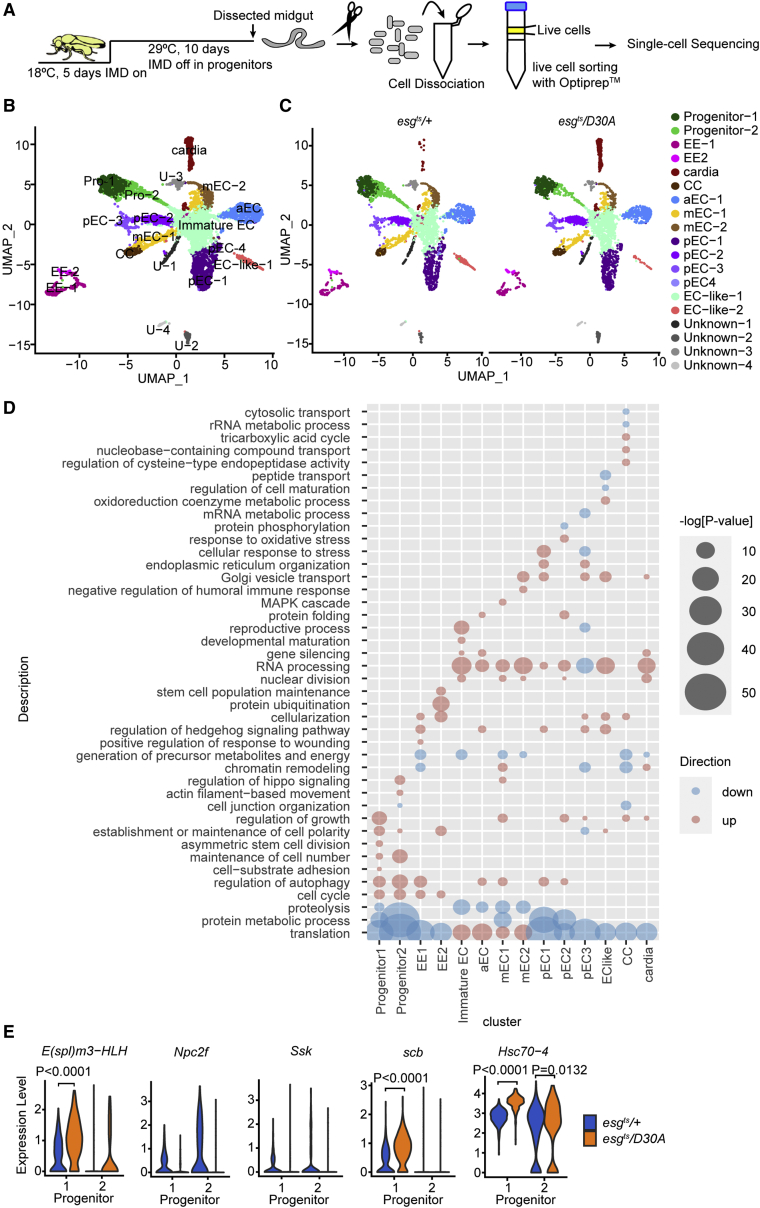
Figure 3.
Inactivating IMD in progenitors affects transcriptional activity in all intestinal epithelial cell types
(A) Schematic representation of an experimental strategy for single-cell transcriptomic analysis of purified intestinal epithelial cells from esgts/+ and esgts/D30A flies.
(B) UMAP plot visualizing cell types in integrated data from esgts/+ and esgts/D30A flies based on the expression of marker genes. Cells are color-coded by cell type.
(C) The same data from (B), split into the labeled genotypes. EE, enteroendocrine cells; CC, copper cells; EC, enterocytes subdivided according to anterior-posterior distribution along the intestine (a, anterior; m, middle; p, posterior).
(D) Gene Ontology term analysis of cell-type-specific processes significantly affected by progenitor-restricted inhibition of IMD. Bubble size indicates the log-transformed significance of the respective enrichments. Pink bubbles indicate enhanced terms, and blue bubbles indicate underrepresented terms.
(E) Representative violin plots of expression levels for the indicated genes in progenitors of esgts/+ and esgts/D30A flies. p values indicate significantly different expression levels. For Npc2f and Snakeskin (Ssk), no expression was observed in progenitors of esgts/D30A flies. See also Figures S3 and S4.