Figure 3.
WGS can help identify specific deleterious on-target effects of CRISPR/Cas9 editing and detect clones falsely identified as corrected
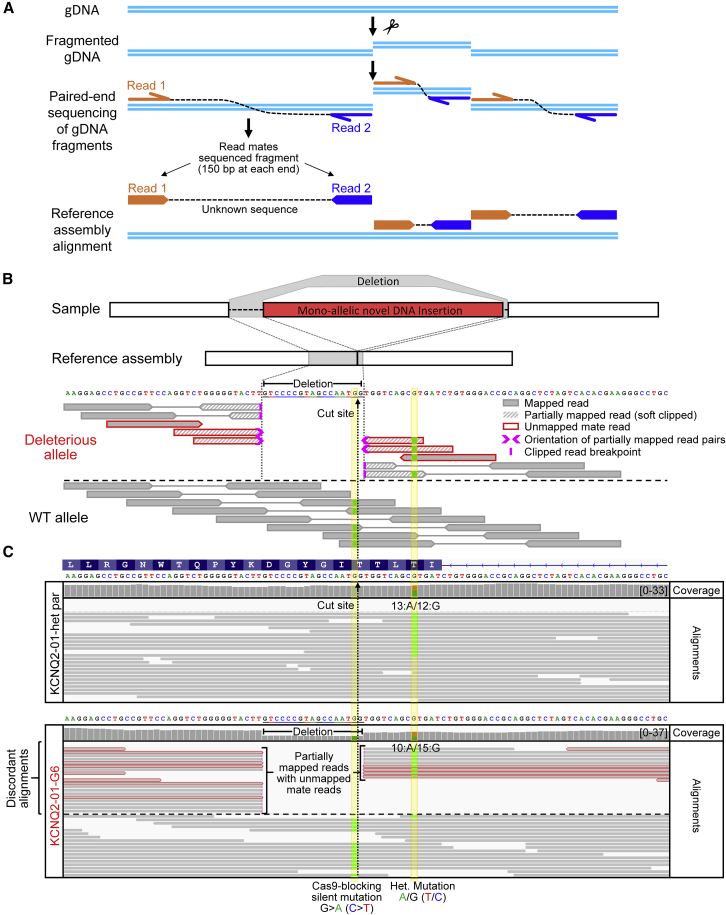
(A) Illustration of paired-end WGS. gDNA is fragmented into random size fragments, and then 150 bp are sequenced from both ends; the insert between the sequenced reads is unknown. Read mate sequences are aligned to a reference genome assembly.
(B) Illustration of paired-end WGS alignments in the presence of a monoallelic structural variant such as the large on-target insertion in KCNQ2-01-G6. A short 17 bp deletion was also introduced around the cut site.
(C) WGS Integrative Genomic Viewer (IGV) plot showing human genome (T2T) reference assembly mapped sequencing reads around the targeted locus of KCNQ2-01-het-parental and -G6 edited line. WGS analysis revealed the presence of the heterozygous patient mutation in both KCNQ2-01-G6 and KCNQ2-01-A6 clones (see Figure S9) that had appeared to be corrected by Sanger sequencing in Figure 1B. When alignments are grouped by concordance to reference assembly, partially mapped reads are displayed at the top. Reads with unmapped mates are outlined in red. Sequence alignment stops abruptly near the Cas9 cut site, with soft clipped bases and unmapped read mate sequences that are homologous to Cas9 carrying plasmid used to target this locus (see also Figure S9).