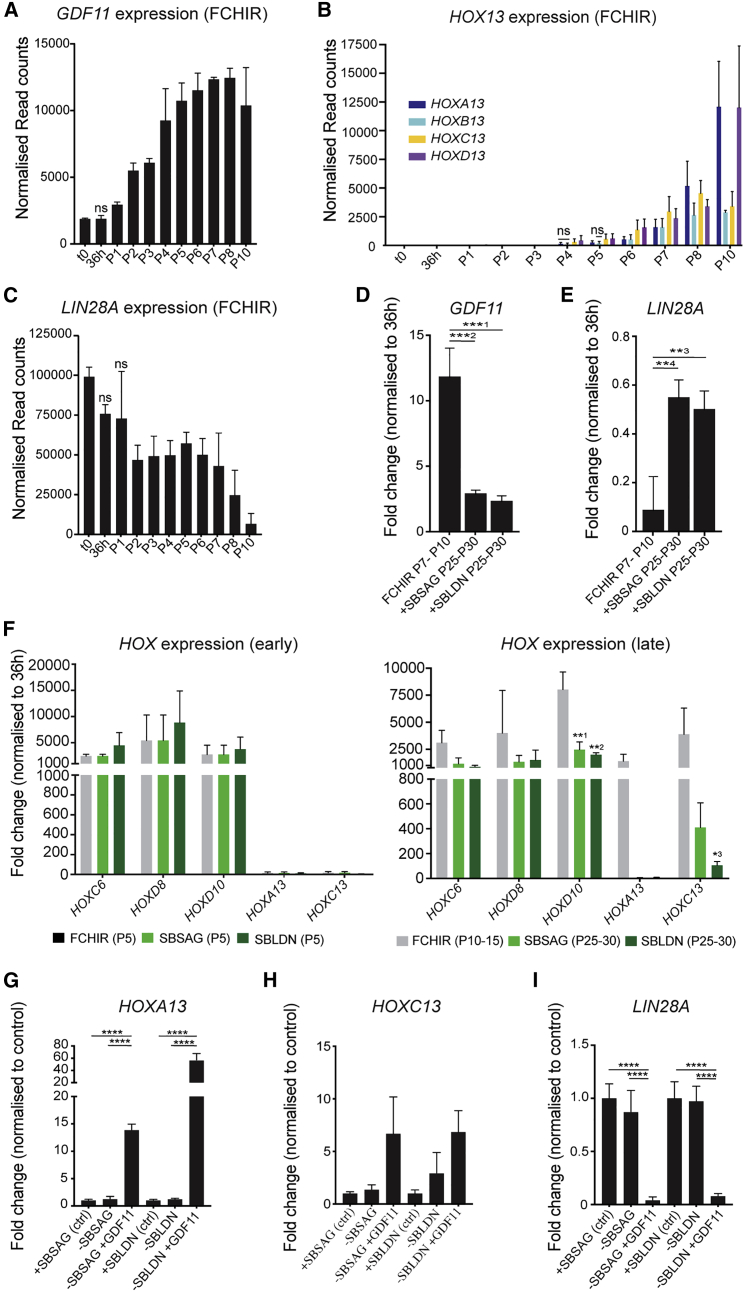
Figure 6.
Modulation of TGF-β and SHH signaling locks in A-P information
(A–C) Normalized expression levels of GDF11 (A), HOX13 (B), and LIN28A (C) at each passage as determined by RNA-seq. Error bars show SEM (n = 3 independent differentiations). All time points were called significantly differentially expressed (FDR < 1%, a FC of at least ±2 compared with t0, and a base mean > 100) unless indicated by “ns.”
(D and E) Transcriptional quantification (qRT-PCR) of GDF11 (D) and LIN28A (E) shown by FC over 36 h and normalized to the reference gene PBGD in late passage PNPs. Error bars show mean with SEM (n = 3 independent differentiations). ∗∗1p = 0.0003, ∗∗2p = 0.0002, ∗∗3p = 0.0026, ∗∗4p = 0.0045 (ANOVA, followed by Fisher’s least significant difference [LSD] multiple comparisons test [MCT]).
(F) Graphs showing the transcriptional quantification (qRT-PCR) of selected HOX genes at early (P5) and late passages (P10–15 or P25–30) in all conditions, ∗∗1p = 0.0039, ∗∗2p = 0.0020, ∗3p = 0.0378 (ANOVA, followed by Fisher’s LSD MCT). Expression levels are presented as FC over the 36 h time point and were normalized to the reference gene PBGD. Error bars show mean with SEM (n = 3 independent differentiations).
(G–I) Transcriptional quantification (qRT-PCR) of HOXA13 (G), HOXC13 (H), and LIN28A (I) in +SBLDN or +SBSAG (ctrl) conditions compared with either +SBLDN or +SBSAG without SB, LDN, or SAG (−SBLDN/−SBSAG) or with GDF11 alone (−SBLDN/−SBSAG + GDF11). Expression levels normalized to the reference gene PBGD. Error bars show SEM (n = 3 independent differentiations). ∗∗∗∗p < 0.001 (ANOVA, followed by Fisher’s LSD MCT).