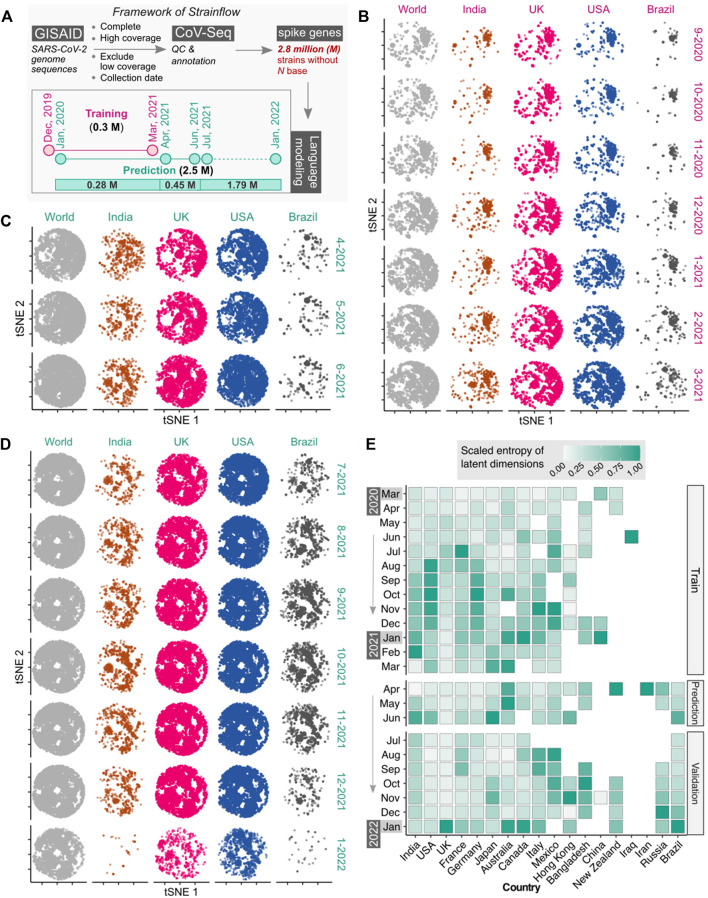
FIGURE 2.
Latent space of spike genes derived using Strainflow preserves spatiotemporal information of SARS-CoV-2 spread. (A) The implementation framework of Strainflow (details described in the method section) (B) tSNE plot showing distinct spatio-temporal relationship based on the latent space learned from the spike gene of 0.308 million SARS-CoV-2 genomes collected till 31 March 2021 (world), India, United Kingdom, United States, and Brazil. (C) Embeddings estimated or predicted from the Strainflow model for 0.45 million SARS-CoV-2 spike genes from the month of April, 2021 to June, 2021. (D) Embeddings estimated or predicted from the Strainflow model for 1.79 million SARS-CoV-2 spike genes from the month of July, 2021 to January, 2022. (E) Heatmap showing the scaled entropy for 18 countries from March, 2020 to January, 2022 (showing data for a. training: March, 2020 to March, 2021, b. prediction: April, 2021 to June, 2021, and c. validation: July, 2021 to January, 2022). The entropies for each country were scaled to the same range to visualize the temporal trends within the country.