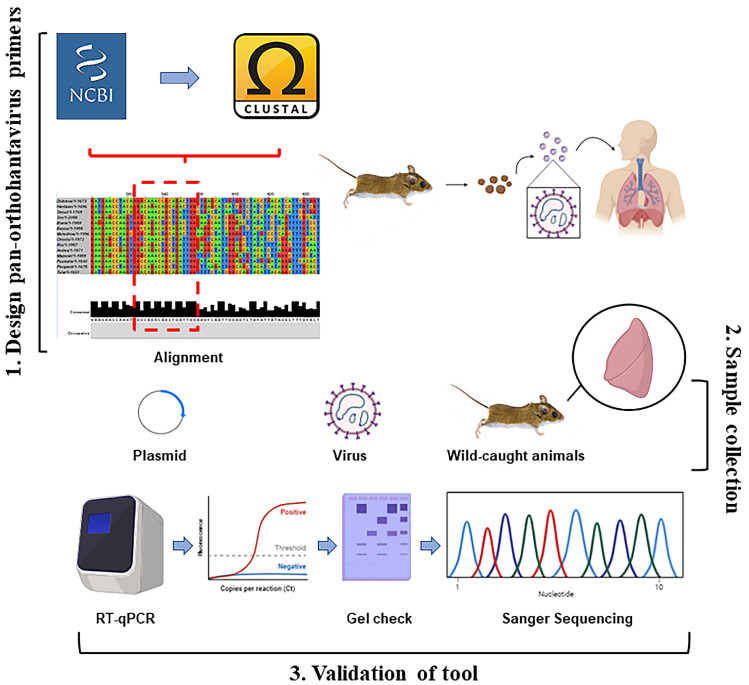
Figure 5.
Overall summary of two-step RT-qPCR pan-orthohantavirus detection tool. 1. S segment sequences from both New and Old World orthohantaviruses were compiled and aligned through NCBI and CLUSTAL Omega. Little to no completely conserved regions existed; therefore, degenerate primers were designed. 2. Samples (plasmids, in vitro cultured virus, and lung tissue from wild-caught rodents) were used to validate primer selection. 3. Two-step RT-qPCR was performed followed by DNA gel electrophoresis. Sanger sequencing was also used for additional confirmation in positive lung tissue samples from wild-caught rodents.