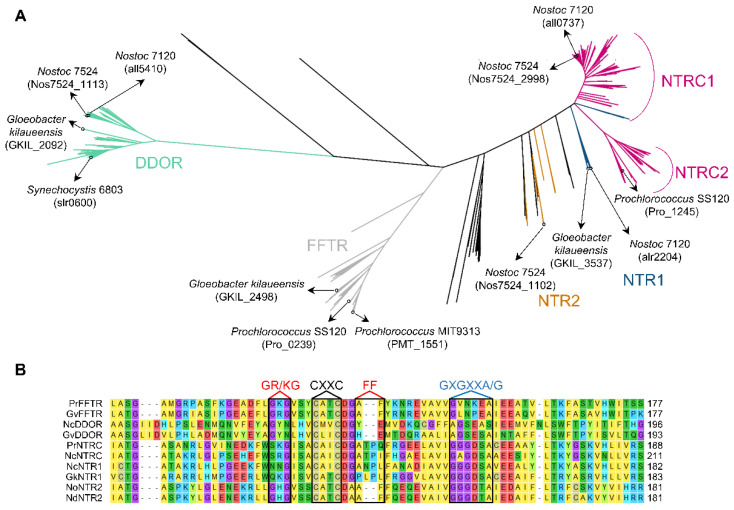
Figure 3.
Functional features and evolutionary relationships of TR-related enzymes in cyanobacteria. (A) Unrooted tree of amino acid sequences of TR-related enzymes identified in the IMG and NCBI databases. Branches are colored to represent proteins found in the different subclades: DDOR (light blue), FFTR (grey), NTRC (purple), NTR1 (dark blue), and NTR2 (brown). Branches related to E. coli, Melainabacteria, Sericytochromatia, and Margulisbacteria are colored black. (B) Multiple protein sequence alignment of several TR-related enzymes. Sequences are indicated with the following abbreviations for organism names: Prochlorococcus marinus SS120 FFTR (PrFFTR), Gloeobacter violaceus FFTR (GvFFTR), Nostoc sp. PCC 7120 DDOR (NcDDOR), Gloeobacter violaceus DDOR (GvDOR), Prochlorococcus marinus SS120 NTRC (PrNTRC), Nostoc sp. PCC 7120 NTRC (NcNTRC), Nostoc sp. PCC 7120 NTR1 (NcNTR1), Gloeobacter kilaueensis NTR1 (GkNTR1), Nostoc sp. PCC 7524 NTR2 (NoNTR2), and Nodularia spumigena CCY9414 (NdNTR2). The motifs for Trx-binding (GR/KG and FF) and NAD(P)H-binding (GXGXXA/G) are shown with boxes in red and blue letters, respectively. Black letters indicate the redox-active Cys (CxxC). The full sequences of each TR-related enzyme are available in Figure S4.