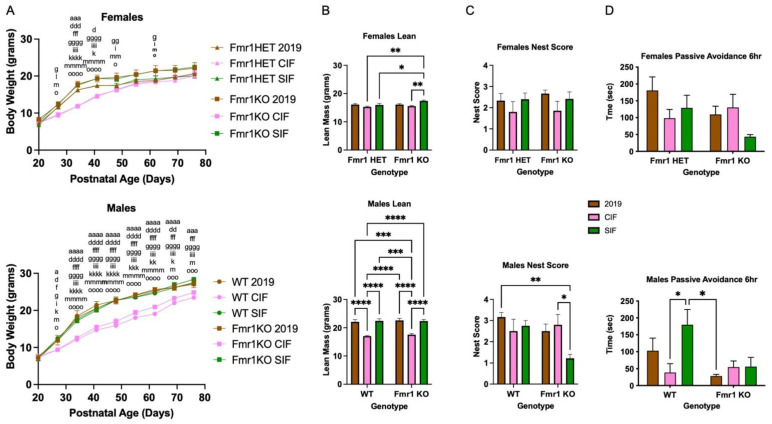
Figure 2.
Body weight and behavior of adolescent WT and Fmr1KO male mice in response to cow milk- and soy-based infant formula diets after 2 months treatment. Fmr1HET females were bred and maintained on Teklad2019 (grain-based but low phytoestrogen levels) diet throughout gestation and lactation. Female Fmr1HET and Fmr1KO and male WT and Fmr1KO pups were weaned at P20 and randomized to diets (Teklad 2019, CIF, SIF). (A) The mice were weighed once per week for 2 months. Female cohorts included Fmr1HET/Teklad 2019 (n = 4), Fmr1HET/CIF (n = 5), Fmr1HET/SIF (n = 6), Fmr1KO/Teklad 2019 (n = 7), Fmr1KO/CIF (n = 7) and Fmr1KO/SIF (n = 6). Statistics were determined by 2-way mixed effects model ANOVA: time F(8230) = 1413, p < 0.0001; genotype/diet F(5,29) = 5.122, p = 0.0017; time x genotype/diet F(40,230) = 8.714, p < 0.0001. Male cohorts included the following: WT/Teklad 2019 (n = 6), WT/CIF (n = 6), WT//SIF (n = 4), Fmr1KO/Teklad 2019 (n = 5), Fmr1KO/CIF (n = 5) and Fmr1KO/SIF (n = 7). Statistics were determined by 2-way mixed-effects model ANOVA: time F(8216) = 2550, p < 0.0001; genotype/diet F(5,27) = 16.78, p < 0.0001; time x genotype/diet F(40,216) = 13.34, p < 0.0001. Tukey’s multiple comparison tests are denoted by p < 0.05 (x), p < 0.01 (xx), p < 0.001 (xxx), and p < 0.0001 (xxxx) where females x = a for Fmr1HET Teklad 2019 versus Fmr1HET CIF, x = b for Fmr1HET Teklad 2019 versus Fmr1HET SIF, x = c for Fmr1HET Teklad 2019 versus Fmr1KO 2019, x = d for Fmr1HET Teklad 2019 versus Fmr1KO CIF, x = e for Fmr1HET Teklad 2019 versus Fmr1KO SIF, x = f for Fmr1HET CIF versus Fmr1HET SIF, x = g for Fmr1HET CIF versus Fmr1KO 2019, x = h for Fmr1HET CIF versus Fmr1KO CIF, x = i for Fmr1HET CIF versus Fmr1KO SIF, x = j for Fmr1HET SIF versus Fmr1KO 2019, x = k for Fmr1HET SIF versus Fmr1KO CIF, x = l for Fmr1HET SIF versus Fmr1KO SIF, x = m for Fmr1KO Teklad 2019 versus Fmr1KO CIF, x = n for Fmr1KO Teklad 2019 versus Fmr1KO SIF, and x = o for Fmr1KO CIF versus Fmr1KO SIF, and males x = a for WT Teklad 2019 versus WT CIF, x = b for WT Teklad 2019 versus WT SIF, x = c for WT Teklad 2019 versus Fmr1KO 2019, x = d for WT Teklad 2019 versus Fmr1KO CIF, x = e for WT Teklad 2019 versus Fmr1KO SIF, x = f for WT CIF versus WT SIF, x = g for WT CIF versus Fmr1KO 2019, x = h for WT CIF versus Fmr1KO CIF, x = i for WT CIF versus Fmr1KO SIF, x = j for WT SIF versus Fmr1KO 2019, x = k for WT SIF versus Fmr1KO CIF, x = l for WT SIF versus Fmr1KO SIF, x = m for Fmr1KO Teklad 2019 versus Fmr1KO CIF, x = n for Fmr1KO Teklad 2019 versus Fmr1KO SIF, and x = o for Fmr1KO CIF versus Fmr1KO SIF. (B) EchoMRI measurement of lean body mass. Female cohorts include Fmr1HET/Teklad 2019 (n = 4), Fmr1HET/CIF (n = 5), Fmr1HET/SIF (n = 6), Fmr1KO/Teklad 2019 (n = 6), Fmr1KO/CIF (n = 7) and Fmr1KO/SIF (n = 6). Statistics were determined by two-way ANOVA: interaction F(2,28) = 2.702, p = 0.085; genotype F(1,28) = 4.410, p = 0.045; diet F(2,28) = 7.438, p = 0.0026. Male cohorts include WT/Teklad 2019 (n = 6), WT/CIF (n = 6), WT/SIF (n = 4), Fmr1KO/Teklad 2019 (n = 6), Fmr1KO/CIF (n = 5) and Fmr1KO/SIF (n = 7). Statistics were determined by two-way ANOVA: interaction F(2,28) = 0.1425, p = 0.87; genotype F(1,28) = 0.3857, p = 0.54; diet F(2,28) = 46.03, p < 0.0001. Tukey’s multiple comparison tests denoted by p < 0.05 (*), p < 0.01 (**), p < 0.001 (***), and p < 0.0001 (****). Total body weight, fat mass, total water mass, and free water mass measurements assessed by EchoMRI are provided in the Supplementary Data. (C) Nest building female cohorts include Fmr1HET/Teklad 2019 (n = 3), Fmr1HET/CIF (n = 5), Fmr1HET/SIF (n = 5), Fmr1KO/Teklad 2019 (n = 6), Fmr1KO/CIF (n = 7) and Fmr1KO/SIF (n = 6). Statistics were determined by two-way ANOVA: interaction F(2,26) = 0.008736, p = 0.92; genotype F(1,26) = 0.1821, p = 0.67; diet F(2,26) = 1.86, p = 0.18. Male cohorts include WT/Teklad 2019 (n = 6), WT/CIF (n = 6), WT/SIF (n = 4), Fmr1KO/Teklad 2019 (n = 6), Fmr1KO/CIF (n = 5) and Fmr1KO/SIF (n = 7). Statistics were determined by two-way ANOVA: interaction F(2,28) = 2.94, p = 0.069; genotype F(1,28) = 4.376, p = 0.046; diet F(2,28) = 2.845, p = 0.075. Tukey’s multiple comparison tests denoted by p < 0.05 (*) and p < 0.01 (**). (D) Passive avoidance female cohorts include Fmr1HET/Teklad 2019 (n = 4), Fmr1HET/CIF (n = 5), Fmr1HET/SIF (n = 6), Fmr1KO/Teklad 2019 (n = 6), Fmr1KO/CIF (n = 7) and Fmr1KO/SIF (n = 6). Statistics were determined by two-way ANOVA: interaction F(2,28) = 2.093, p = 0.14; genotype F(1,28) = 2.530, p = 0.12; diet F(2,28) = 1.642, p = 0.21. Male cohorts include WT/Teklad 2019 (n = 6), WT/CIF (n = 6), WT/SIF (n = 4), Fmr1KO/Teklad 2019 (n = 6), Fmr1KO/CIF (n = 5) and Fmr1KO/SIF (n = 7). Statistics were determined by two-way ANOVA: interaction F(2,28) = 2.998, p = 0.066; genotype F(1,28) = 6.844, p = 0.014; diet F(2,28) = 3.229, p = 0.055. Tukey’s multiple comparison tests denoted by p < 0.05 (*). Data shown for 6-hours post-foot shock. Results were not statistically different for 24- and 48-h post-foot shock.