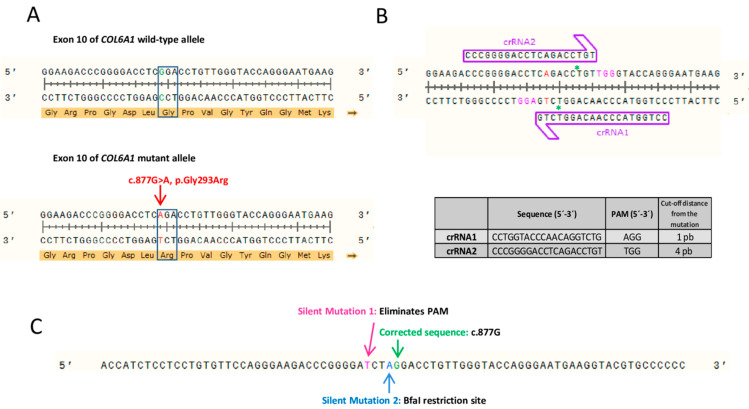
Figure 1.
Strategy for CRISPR/Cas9 editing of c.877G>A variant in exon 10 of COL6A1. (A) The c.877G>A, p.Gly293Arg dominant-negative pathogenic variant is heterozygous in the cohort of individuals studied here. (B) Schematic representation of the experimental design of the two RNA guides (crRNA1 and crRNA2). Mutant adenine is shown in red. Protospacer adjacent motif (PAM) sequences are highlighted in pink for each crRNA. Green asterisks (*) represent the predicted Cas9 cleavage site for each crRNA. The table shows the sequence of each of the crRNAs, their corresponding PAM and the nucleotides that separate the Cas9 cut-off point of the variant. (C) Sequence of the single-stranded DNA template delivered to fibroblasts to correct the pathogenic variant by homology-directed repair. In addition to the wild-type guanine nucleotide at position c.877, it contains two extra silent changes to eliminate the PAM and generate a restriction site for the BfaI.