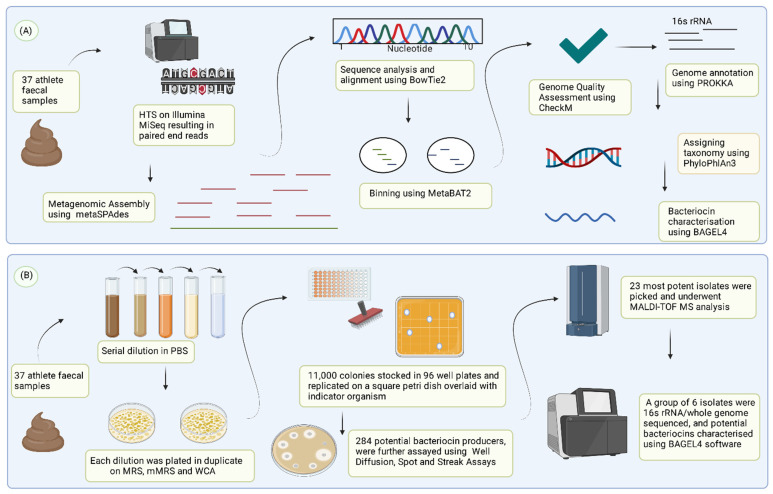
Figure 1.
In silico and in vitro based approaches used in this study to identify potential novel bacteriocins from the athlete’s gut. (A) Metagenomic data from 37 faecal samples in the form of paired-end reads were assembled, annotated, quality-checked, and binned to recover Metagenome-Assembled Genomes (MAGS) analysed using BAGEL4 for the presence of potential bacteriocin genes. (B) 37 faecal samples from elite Irish athletes were screened for novel bacteriocin-producing gut isolates. Potential bacteriocin producers were assayed further, and the spectrum of inhibition was assessed. Isolates exhibiting potential antimicrobial activity were brought forward for MALDI-TOF mass spectrophotometry, whole-genome sequencing (WGS), and bacteriocin biosynthetic gene clusters were predicted using BAGEL4 software. (Figure created with https://BioRender.com, accessed on 15 February 2022).