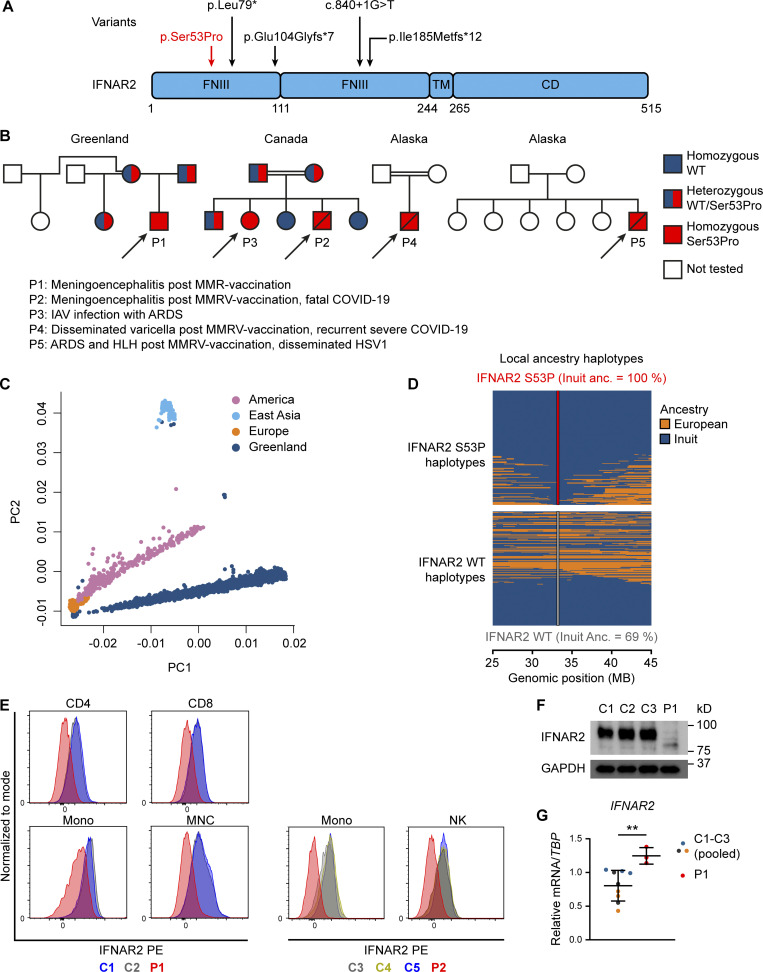
Figure 1.
Identification of a novel homozygous IFNAR2 variant leading to IFNAR2 deficiency. (A) IFNAR2 protein sequence with known deleterious variants marked. (B) Pedigrees of affected individuals originating from Greenland (P1), Canada (P2 and P3), and Alaska (P4 and P5). Double lines refer to consanguinity. (C) Principal component analysis of 4,630 Greenlandic individuals combined with three super populations from the 1000 Genomes Project. (D) Local ancestry haplotypes of admixed and p.Ser53Pro IFNAR2 heterozygous Greenlandic individuals colored according to ancestry. Each heterozygous individual is represented with two haplotypes, one for the p.Ser53Pro haplotype (top) and one for the WT haplotype (bottom). The fraction of Inuit ancestry of the WT and p.Ser53Pro haplotype is estimated at the location of the variant. (E) IFNAR2 cell surface expression analyzed by flow cytometry in PBMCs and distinct cell subsets of healthy controls (C1–C5) and patients (P1 and P2). Representative of a single experiment in P1 and P2 PBMC. (F) Immunoblotting showing IFNAR2 expression in whole-cell lysates of PBMCs from P1 compared to three healthy controls (C1–C3). GAPDH was used as loading control. Not repeated due to limited patient material. (G) RT-qPCR of whole-cell RNA lysates of PBMCs evaluating IFNAR2 mRNA levels relative to TBP for P1 compared to three healthy controls (pooled, mean ± SD of n = 3 biological replicates; **, P < 0.01, Welch’s t test). Representative of a single experiment in P1 PBMC. Source data are available for this figure: SourceData F1.