Fig. 1.
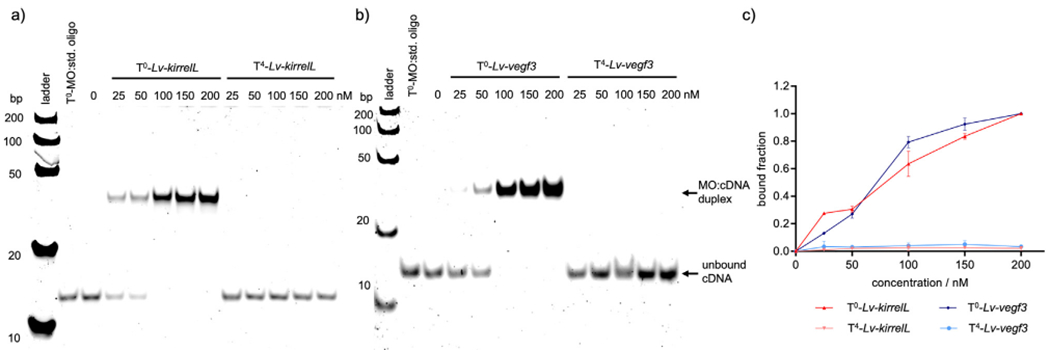
Representative gel shift assays of non-caged (T0) and caged (T4) MOs interacting with their corresponding complementary target DNA (100 nM) sequences. a) T0-Lv-kirrelL and T4-Lv-kirrelL, b) T0- Lv-vegf3 and T4- Lv-vegf3 and c) quantification of the gel shift assay. A Fast Ruler Ultra Low Range ds-DNA Ladder (SM1233, ThermoFisher Scientific) was used. A standard control oligo from Gene Tools (denoted as “std. oligo”) was incubated with T0-MOs in equimolar amounts as a negative control. Gel images were quantified with ImageLab, and the bound MO:cDNA fraction was plotted against MO concentration. Error bars represent the standard deviation of three independent trials.
