Abstract
Background
Multidrug resistance (MDR) mediated by ATP binding cassette subfamily B member 1 (ABCB1/P-gp) is a major cause of cancer chemotherapy failure, but the regulation mechanisms are largely unknown.
Methods
Based on single gene knockout, we studied the regulation of CDK6-PI3K axis on ABCB1-mediated MDR in human cancer cells. CRISPR/Cas9 technique was performed in KB-C2 cells to knockout cdk6 or cdk4 gene. Western blot, RT-PCR and transcriptome analysis were performed to investigate target gene deletion and expression of critical signaling factors. The effect of cdk4 or cdk6 deficiency on cell apoptosis and the cell cycle was analyzed using flow cytometry. In vivo studies were performed to study the sensitivity of KB-C2 tumors to doxorubicin, tumor growth and metastasis.
Results
Deficiency of cdk6 led to remarkable downregulation of ABCB1 expression and reversal of ABCB1-mediated MDR. Transcriptomic analysis revealed that CDK6 knockout regulated a series of signaling factors, among them, PI3K 110α and 110β, KRAS and MAPK10 were downregulated, and FOS-promoting cell autophagy and CXCL1-regulating multiple factors were upregulated. Notably, PI3K 110α/110β deficiency in-return downregulated CDK6 and the CDK6-PI3K axis synergizes in regulating ABCB1 expression, which strengthened the regulation of ABCB1 over single regulation by either CDK6 or PI3K 110α/110β. High frequency of alternative splicing (AS) of premature ABCB1 mRNA induced by CDK6, CDK4 or PI3K 110α/110β level change was confirmed to alter the ABCB1 level, among them 10 common skipped exon (SE) events were found. In vivo experiments demonstrated that loss of cdk6 remarkably increased the sensitivity of KB-C2 tumors to doxorubicin by increasing drug accumulation of the tumors, resulting in remarkable inhibition of tumor growth and metastasis, as well as KB-C2 survival in the nude mice.
Conclusions
CDK6-PI3K as a new target signaling axis to reverse ABCB1-mediated MDR is reported for the first time in cancers. Pathways leading to inhibition of cancer cell proliferation were revealed to be accompanied by CDK6 deficiency.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12943-022-01524-w.
Keywords: Multidrug resistance (MDR), cancer, Transcriptome sequencing, Cyclin dependent kinase 6 (CDK6), PI3K 110α/110β, Signaling axis, ATP-binding cassette (ABC) transporter ABCB1/P-gp
Introduction
Multidrug resistance (MDR) can develop in cancer cells that survive during chemotherapy. P-glycoprotein (P-gp), revealed as ATP-binding cassette (ABC) subfamily B member 1 (ABCB1) is an ABC transporter and its overexpression by various cancers can produce resistance to various chemotherapeutic drugs that have distinct structures and differing mechanisms of action [1], which hurdles the efficiency of chemotherapy against cancers. Despite the reversal efficacy of various types of ABCB1 inhibitors, mechanisms of inducing and regulating ABCB1-mediated MDR are rarely reported on the level of signaling pathways. Lack of understanding of ABCB1 regulation may lead to increased risk of side effects when treated with inhibitors, most of which are due to the lack of specificity.
An ideal target can exert its efficiency to inhibit cancer cell proliferation when it is used for attenuating MDR in cancer. Through CRISPR/Cas9 gene editing, we recently reported that 110α and 110β subunits of the phosphoinositide 3-kinase (PI3K) signaling pathway are potential targets for reversal of ABCB1/P-gp- and ABCG2/BCRP-mediated cancer MDR in a manner independent of AKT expression [2]. With the aim to characterize other efficient and safe targets that can be used to reverse ABCB1-mediated MDR in cancers, we further investigated the signaling pathways involved in the PI3K110α and 110β regulated ABCB1 expression.
The cyclin-dependent kinases (CDKs) are members of a family of serine-threonine kinases that regulate the cell cycle by altering cell proliferation and apoptosis [3–12]. Although many functional studies were based on the inhibitor-triggered variation of protein expression, which were frequently influenced by the cytotoxicity or non-specificity of the inhibitors instead of inhibition of CDKs themselves, some functions are demonstrated by protein-protein interactions. CDKs comprise three domains: the ATP binding and catalytic domain, the cyclin-binding domain, and the P13suc1 binding domain, in which P13suc1 can inhibit the activity of CDK and cell phase alteration [13–15]. By mediating a common kinase domain containing peptide, PSTAIRE, CDKs bind to their corresponding cyclin, forming a cyclin-CDK complex, where CDKs catalyze the phosphorylation of certain serine and threonine residues in target proteins. This regulates gene transcription and cell division, further promoting alteration of cell cycle between different phases, e.g., phase G1 to S, G2 to M, and exit from phase M [16].
CDK4 and CDK6 are essential kinases and are drawing increasing attention because they can drive cell proliferation by combining with cyclin D1, D2, and D3 [17]. Imbalance in the activity of CDK4 and CDK6 can induce dysregulation and uncontrolled cell division, which is a hallmark of cancers [17]. The interaction of CDK4 and CDK6 with D-type cyclins constitutively phosphorylates and inactivates the retinoblastoma protein (Rb), a tumor suppressor, causing the release of its binding partner, E2 transcription factor (E2F), allowing cancer cells to overcome pRb-dependent growth suppression [18]. Furthermore, CDK4 and CDK6 can bind to the proteins p15, p16, p21 and p27 [19, 20], and are regulated by cytosolic sialidase (Neu2) [21], FAT1 [22], SRY homology box 2 (SOX2) [23], reactive oxygen species (ROS) [24], and more. In addition, CDK6 may regulate epithelial-mesenchymal transition (EMT) via the CCNA2/MET pathway [25]. CDK6 also regulates malignant stem cell quiescence and facilitates nuclear factor kappaB (NF-κB) signaling [26]. The downregulation or inhibition of CDK6 or CDK 4 induces cellular apoptosis, suppresses tumor proliferation, migration, and invasion [27, 28].
Currently, whether CDK4 and/or CDK6 have direct regulatory effects on ABCB1 and ABCB1-mediated MDR in cancer, their roles, and mechanisms are unknown. Most of the inhibitors lack specificity, and some of them cannot accumulate inside the cells efficiently because they may be substrates of ABCB1 [29–31]. In this study, by gene knockout with CRISPR/Cas9 gene editing technique, we determined the regulation of CDK4 or CDK6 on ABCB1-mediated MDR in human epidermoid carcinoma MDR cell line KB-C2 which overexpresses ABCB1 and H460/MX80 which expresses ABCB1 at low level. Then we focused on the coordination of CDK6 with PI3K 110α/β to regulate ABCB1 expression, and their function to induce alternative splicing (AS) of ABCB1 pre-mRNA. Importantly, this study demonstrated that CDK6 is a novel multi-functional target that may act as an on-off switch for ABCB1 expression, potentially reversing ABCB1-mediated MDR in cancers, and therefore, has significance in combination with cancer chemotherapy.
Materials and methods
Cells, plasmids, and chemicals
The human epidermoid carcinoma MDR cell line KB-C2 created by exposing KB-3-1 cells to increasing concentrations of colchicine and its parental cell line KB-3-1, which were used as a pair of models for studying ABCB1-mediated MDR in cancer [32], were kindly provided by Dr. Shin-ichi Akiyama (Kagoshima University, Kagoshima, Japan). Non-small cell lung cancer (NSCLC) NCl-H460 cell line was kindly provided by Drs. Susan E. Bates and Robert W. Robey (NIH, Bethesda, MD). H460/MX80 cells with enhanced drug-resistant ability were generated in our previous study [2]. KB-C2-k.o.110α, KB-C2-k.o.110β, MX80-k.o.110α, or MX80-k.o.110β gene-deficient cell populations were constructed using CRISPR/Cas9 editing technology according to our previous study [2]. The cells were cultivated with DMEM supplemented with 10% FBS and 1% penicillin/streptomycin in a humidified incubator containing 5% CO2 at 37 °C. CRISPR/Cas9 all-in-one plasmids, encoding single guide RNA (SgRNA) and Cas9, were purchased from GeneCopoeia Inc. (Rockville, MD).
Ribociclib was kindly provided by ChemieTek (Indianapolis, IN). Paclitaxel, colchicine, and DOX were purchased form Sigma Chemical Co. (St. Louis, MO). Mouse anti-ABCB1, HRP ligated or fluorescent secondary rabbit or goat-anti mouse antibodies, were purchased from Invitrogen, Thermo Fisher (Carlsbad, CA). Mouse-anti-CDK4 and CDK6 antibodies were purchased from R&D Systems (Minneapolis, MN). Rabbit-anti-CKAP4 antibody and Cy3- or FITC-labeled goat-anti-rabbit antibodies were purchased from Servicebio technology co., LTD (Wuhan, China). The other reagents were purchased from VWR International (West Chester, PA).
Determination of cell viability: MTT assay
Exponentially growing cells were seeded into 96 well plates at 5 × 103 cells/well. After 72 h of incubation, 20 μL of MTT (5 mg/mL) was added to each well. After incubation for an additional 4 h, the medium containing MTT was discarded and replaced with 150 μL of DMSO to dissolve the dark blue-purple crystal. The absorbance was measured at a wavelength of 490 nm, using an ELx 800 Universal Microplate Reader (Bio-Tek, Inc. Winooski, VT). The relative survival rate for the cells was analyzed using the SPSS 20 program (SPSS Inc., Chicago, IL) and the survival rate – drug concentration curves were generated using Origin 9.0 software (OriginLab Corporation, Northampton, MA). The concentration of drug required to inhibit cell viability by 50% (IC50 value) was determined using Origin 9.0 software.
Western blot assay and immunofluorescence (IF) analysis
Parental KB-3-1 cells and MDR KB-C2 cells were incubated with 9 μM of ribociclib for 2 h and co-cultured with paclitaxel or colchicine for 24 to 72 h. For the Western blot assay, the cells were lysed with SDS lysate reagent, separated on a gradient polyacrylamide gel and transferred to a PVDF membrane. After blocking with 5% milk and washed with TBST buffer, the membrane was incubated with mouse anti-ABCB1 antibody at 4 °C for 2 h, adequately washed with TBST, and incubated with goat anti-mouse IgG-HRP at RT for 2 h. The membrane was then washed with TBST and exposed to the SignalFire™ ECL Reagent developing reagent (Cell Singling Technology, Danvers, MA), and the results were quantified using a AI600 RGB GEL Imaging System (GE, Fairfield, CT) set for the chemiluminescence mode.
For the IF analysis, KB-C2 and KB-3-1 cells were washed with PBS buffer (pH 8.0), fixed with 4% formaldehyde, followed by incubation with 0.1% of Triton-100 and then 6% BSA. The cells were co-incubated with ABCB1-specific antibodies (mouse originated, labeled with Alexa Fluor 488 (AF488); Santa Cruz Biotechnology, Inc. CA) for 1 h at 37 °C, washed with PBS for four times and stained with 0.5 μg mL− 1 of 4′,6-diamidino-2-phenylindole (DAPI) in PBS. Fluorescence was determined using a with Cytell™ Image Cytometer (GE Healthcare, Washington).
Analysis of regulation of CDK6-PI3K axis on ABCB1 mediated drug resistance
CRISPR/Cas9 technique was performed in KB-C2 cells to knockout cdk6 or cdk4 gene. Western blot, RT-PCR and transcriptome analysis were performed to investigate target gene deletion and expression of critical signaling factors. Detailed procedure and other methods were described in supplemental materials and methods.
CRISPR/Cas9 knockout and characterization for target gene deficiency
The cells were seeded into 48-well plates with serum-free DMEM. A polymeric GenePORTER transfection reagent was mixed with sgRNA/Cas9 all-in-one expression plasmid HCP254656-CG12–1-10 or HCP288574-CG12–1-10 to target AGTGTGAGAGTCCCCAATGG in cdk4 (NM_000075.3) and GAAGAACGGAGGCCGTTTCG in cdk6 (NM_001259.7), respectively (GeneCopoeia Inc., Rockville, MD). After incubation at room temperature for 10 min, the mixture was added to the cells. After incubation for 4 h, FBS was added to the cultures at 20% of the final volume ratio. To achieve a high transfection ratio, the same well of cells were transfected again after 48 h of cultivation. The drugs used in this study were not used during transfection and cell stabilization, and RT-PCR and Western blotting were performed to determine the transcription and expression of the target genes after stabilizing the gene deficient cells for 1 month via continuous cell cultivation.
To determine the transcripts for cdk4, primers (5′) GCTACCTCTCGATATG (3′) and (5′) AGCCTGGTGGGGGTGC (3′) were used for RT-PCR and for cdk6, the primers (5′) GAGAAGGACGGCCTGT (3′) and (5′) TCAAGTCTTGATCGAC (3′) were used.
Analysis of CDK4 or CDK6 to regulate ABCB1 expression and ABCB1-mediated anti-drug efficacy
In the KB-C2 cells, either the cdk4 or cdk6 gene was deleted and the cells were cultured for 48 h and lysed with SDS lysate reagent. Western blot analysis of cell lysate was performed as described above for the determination of the effect of cdk4 or cdk6 knockout on the expression of the ABCB1 protein. The gene deficient cells were cultured with gradient concentrations (0.001, 0.01, 0.1, 1, 10 or 100 μM) of either colchicine or paclitaxel for 72 h. The MTT assay and IC50 determination was carried out as described above.
Differential expression analysis and identification of AS events
Cells were lysed and Total RNA was extracted using the TRIzol® Reagent, according to the manufacturer’s instructions (Invitrogen). Genomic DNA was then removed using Dnase I (Takara Bio, Otsu, Japan). RNA quality was determined using a 2100 Bioanalyser (Agilent Technologies, Palo Alto, CA, USA) and quantified using ND-2000 (NanoDrop Technologies, Inc. Wilmington, DE, USA). RNA samples (5 μg) were used to construct the RNA-seq transcriptome library. The messenger RNA (mRNA) was fragmentated using a fragmentation buffer and isolated by the polyA selection method using oligo (dT) beads. According to Illumina’s library construction protocol, cDNA originated double-strands DNA (dscDNA) were synthesized, subjected to end-repair, phosphorylation and ‘A’ base addition, selected for cDNA target fragments of 200–300 bp, followed by PCR amplification using Phusion DNA polymerase (NEB) for 15 PCR cycles, and then sequenced using the Illumina HiSeq 4000 (2 × 150 bp read length). After map reading [33], differential expression genes (DEG) between two different samples were identified according to the fragments per kilobase of exon per million mapped reads (FRKM) method [34]. Network of gene clusters and protein-protein interaction were analyzed on metascape (http://metascape.org) [35] or STRING (http://string-db.org) platform, choosing Homo Sapiens as the control species. The data was visualized by drawing heatmaps using TBtools (https://github.com/CJ-Chen/TBtools). For the identification of AS events. All of the AS events that occurred in the samples were identified by using the program, Multivariate Analysis of Transcript Splicing (MATS, http://rnaseq-mats.sourceforge.net/) [36].
Analysis of cell apoptosis using flow cytometry
The KB-C2 cells and the derivative cdk4- or cdk6-deficient cells were seeded into a 24-well plate at 1 × 105 cells per mL of medium and cultured with colchicine (1.25 μM) for 24 h. The cells were treated with 0.25% trypsin, followed by DMEM to terminate trypsin activity and washed two times with cold PBS. The cells incubated with the medium were used as a control group. Cell apoptosis was determined using an Annexin V-FITC Apoptosis Detection Kit (Solarbio Science & Technology Co., Ltd., Beijing, China), following the manufacturer’s instructions. The cellular apoptosis and necrosis ratios were analyzed using a CytoFLEX flow cytometer (Beckman Coulter, Brea, CA).
Determining the effect of cdk4 or cdk6 deficiency on the cell cycle
The KB-C2 cells and cdk4- or cdk6-deficient cells derived from KB-C2 were seeded into a 24-well plate at 1 × 105 cells per mL of medium and cultured with colchicine (1.25 μM) for 24 h. Next, the cells were treated with 0.25% trypsin, followed by DMEM and washed two times with PBS. The cells incubated with medium were used as a control group. The stage of the cell cycle was determined using a Cell Cycle Staining Kit (Multi Sciences (LianKe) Biotech, Co., Ltd., Hangzhou, China), following the manufacturer’s instructions. The cell populations were analyzed using a CytoFLEX flow cytometer (Beckman Coulter, Brea, CA) and the population range corresponding to the untreated KB-C2 cells in the best status (approximately 65% among all cell events), was selected as the gate for all the tested cell groups. All experiments were repeated in triplicate.
In vivo studies
We established two tumor-bearing mouse models to evaluate the reversal of MDR tumor effect of cdk6 deficiency in vivo. To establish the KB-C2 and KB-C2-k.o.cdk6 tumor-bearing mouse models, the aliquots (0.1 mL) of cancer cells in sterilized saline solution (1.0 × 107 cells/mL) were subcutaneously inoculated into the back neck of the nude mice (BALB/c-nu). The mice were randomly divided into 4 groups (9 mice per group) with equivalent average starting tumor size (~ 50 mm3) and body weight (~ 23 g) treated with 0.9% saline (set as control) or DOX (2 mg Kg− 1), via the intraperitoneal route of administration for each tumor-bearing mouse model. Animals were examined daily, and their body weights and tumor sizes were determined using the formula: Tumor size = Length×Width×Height×π/6 [37]. The increased tumor sizes were calculated by subtracting the tumor sizes in the initial group from that of saline- or DOX-treated group at the subsequent measurement time. Animals were sacrificed 19 days after treatment.
The pathological morphology of liver, lung, spleen, heart, stomach and kidney of nude mice after drug withdrawal was analyzed by HE staining. Based on transcriptome analysis, genes with high level of expression in both KB-C2 and KB-C2-k.o.cdk6 are screened. Among them, cytoskeleton-associated protein 4 (CKAP4) was used as a biomarker for determination of distribution of KB-C2 or KB-C2-k.o.cdk6 in different organs including liver, lung and spleen. Immunofluorescence staining was then performed to analyze tumor cell migration in each group of tumor-bearing nude mice.
Statistical analysis
All experiments were performed three times and the results were analyzed using Student’s unpaired t-test using SPSS 20.0 software (SPSS Inc., Chicago, IL). Data were expressed as the mean ± standard error of the mean (SEM). Results showing P values less than 0.05 were considered statistically significant.
Experimental procedures
Reversal of MDR in cancer cells was studied by MTT assay. The expression level of ABCB1 was analyzed by Western blot assay and immunofluorescence (IF). the transcription and expression of the target genes were determined by RT-PCR and Western blotting, respectively, after stabilizing the gene deficient cells for 1 month via continuous cell cultivation. Up- or down- regulation of relative proteins and signaling pathways was studied by differential expression analysis and the mechanism was explored by identification of AS events and protein-protein interaction (PPI). Cell apoptosis and the cell cycle were analyzed by flow cytometry.
Results
Ribociclib, an inhibitor for CDK4/6, could downregulate ABCB1 expression
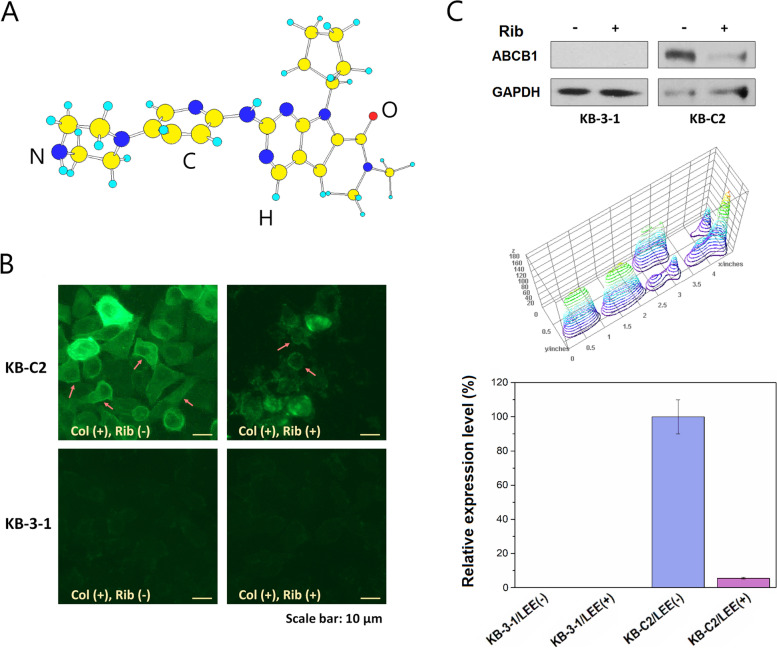
During screening the agents for reversal of ABCB1-mediated MDR, we found that ribociclib (LEE011), an inhibitor for CDK4/6, could significantly downregulate ABCB1 expression in MDR KB-C2 cells, as shown by immunofluorescence results and Western blot (Fig. 1). To our knowledge, most of the CDK4/6 inhibitors did not show the capability to reverse ABCB1-mediated MDR or downregulate ABCB1 expression. This could possibly be related to the low efficacy of CDK4 or CDK6 to regulate ABCB1 expression, or to the low intracellular accumulation efficiency and relatively poor specificity of many known inhibitors. Therefore, it is necessary to perform gene knockout to elucidate whether CDK4 or CDK6 deficiency has a role in reversing ABCB1-mediated drug resistance.
Fig. 1.
Downregulation of ABCB1 expression by ribociclib. A Molecular structure (3-dimentional) of ribociclib. B, C Immunofluorescence (IF) and Western blot data indicating ABCB1 expression was significantly downregulated by incubating the KB-C2 cells with 9 μM of ribociclib (Rib)/LEE011 for 72 h. Before the analysis, KB-C2 and KB-3-1 cells were incubated with 0.07 or 0.01 μM (lower than the IC50 values) of colchicine, respectively (to simulate the conditions with presence of colchicine during reversal of MDR and to maintain normal morphology in cells). ABCB1 expressed on the cell membrane is depicted by arrows in red
The knockout of cdk4 and cdk6 genes using the CRISPR/Cas9 gene editing technique
To explore the potential mechanism of CDK4 or CDK6 in regulating ABCB1 expression, cdk4 and cdk6 genes were knocked out by transfecting KB-C2 cancer cells with the all-in-one CRISPR/Cas9 plasmid harboring the sgRNA that targets either the cdk4 or cdk6 genes (Fig. 2). The transfected cells were stabilized through continuous culture for at least 2 weeks. The deletions of cdk4 or cdk6 genes in KB-C2 produced a significant decrease in the expression of the CDK4 and CDK6 proteins, respectively (Fig. 2B). Western blot analysis (Fig. 2B), RT-PCR analysis (Fig. 2B) and transcriptome sequencing and quantification (Table S1) indicated that cdk4 or cdk6 gene were deleted by chromosomal recombination. The cdk4 or cdk6 deficient KB-C2 cells were named as KB-C2-k.o.cdk4 or KB-C2-k.o.cdk6, respectively.
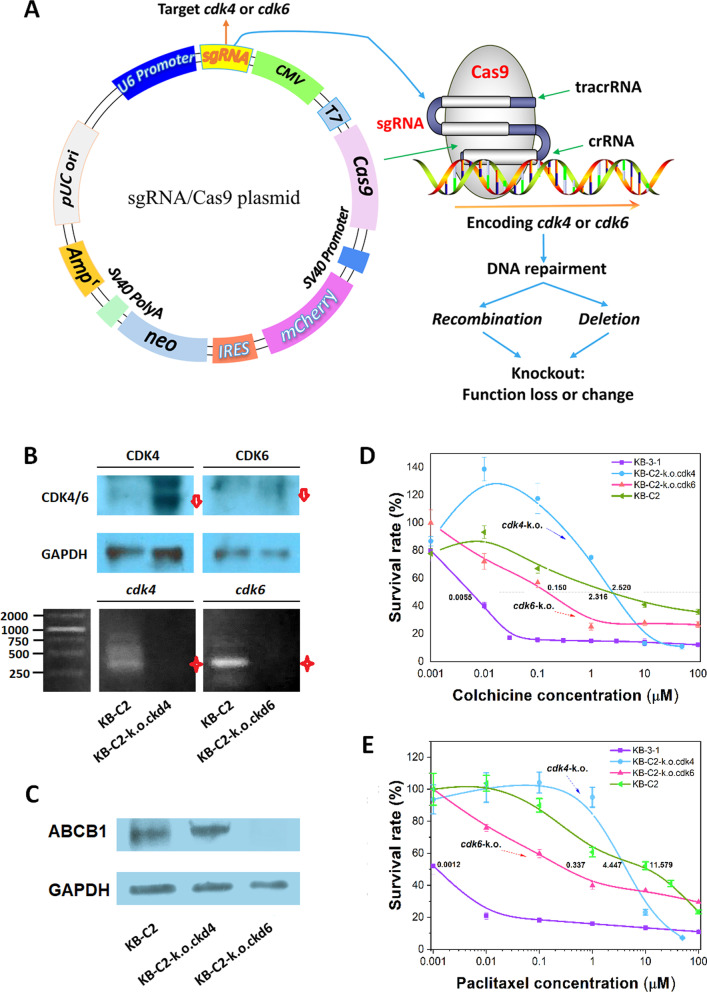
Fig. 2.
The effect of the deletion of cdk4 or cdk6 gene on the regulation of ABCB1 and the reversal of ABCB1-mediated MDR. A An all-in-one plasmid for CRISPR/Cas9 gene editing. B Western blot and RT-PCR analysis verifying the specific knockout of the targeted gene. The red stars depict the decrease in the copies of the target PCR. Truncated proteins are depicted by head-down arrows. C ABCB1 expression in the cdk4- or cdk6-deficient KB-C2 cells. The cell lysates containing identical amounts of total proteins were used for determining gene knockout in (B) and ABCB1 expression (C). D, E The alteration of ABCB1-mediated MDR in the cdk6- or cdk4-deficient KB-C2 cells. Two substrate anticancer drugs of ABCB1, colchicine and paclitaxel, were used to compare ABCB1-mediated MDR in the cancer cells
CDK6 is a potential target for on-off regulation of ABCB1 expression and ABCB1-mediated MDR
Amazingly, the gene deletion experiments indicated that the cdk4 and cdk6 genes have different roles in regulating ABCB1 expression and ABCB1-mediated MDR in KB-C2 cells. Compared with the KB-C2 cells, the deletion of the cdk6 alone induced significant downregulation in the expression of the ABCB1 transporter in the KB-C2-k.o.cdk6 cells (Fig. 2C). The Western blot indicated that wild-type ABCB1 was remarkably downregulated in KB-C2-k.o.cdk6 cells (Fig. 2C) whereas trace amounts of ABCB1 homologous protein with a higher molecular weight might exist. Due to the decreased ABCB1 transporter expression, cdk6-deficient KB-C2 cells achieved remarkable reversal of MDR, showing significant increase in sensitivity to ABCB1 substrates colchicine (Fig. 2D) and paclitaxel (Fig. 2E), demonstrating that CDK6 regulates cancer MDR mediated by ABCB1 specifically.
Similarly, ABCB1 positive protein signal in Western blot was upregulated in KB-C2-k.o.cdk4 population, in which cdk4 gene was knocked out (Fig. 2C). In line with this phenomenon, MDR was increased in the KB-C2-k.o.cdk4 (cdk4 deficient KB-C2) cells incubated with low-to-medium concentrations of colchicine (< 2.5 μM) or paclitaxel (< 4 μM). However, MDR was not enhanced at higher concentrations of these drugs, indicating that KB-C2 cells deficient in CDK4 were more susceptible when incubated with higher concentrations of the drugs. Thus, our results clearly indicate that CDK6 and CDK4 function as possible on-off switches that regulate the expression of the ABCB1-mediated MDR, although CDK4 is homologous to CDK6. Native CDK6 may have a role in maintaining ABCB1-mediated MDR of cancers.
Determining potential pathway(s) for CDK6 to regulate ABCB1 expression
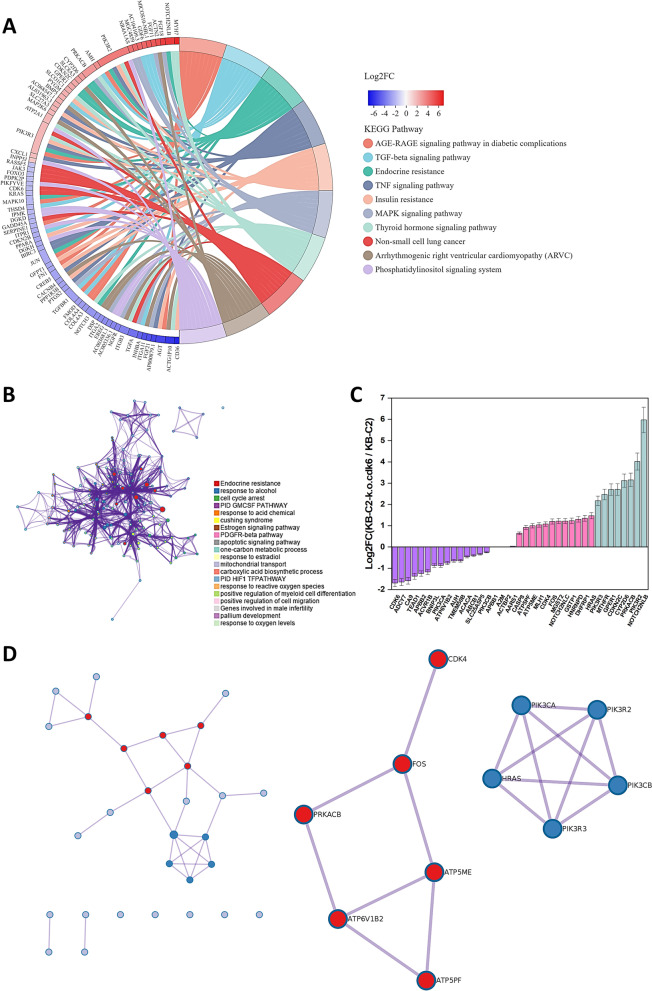
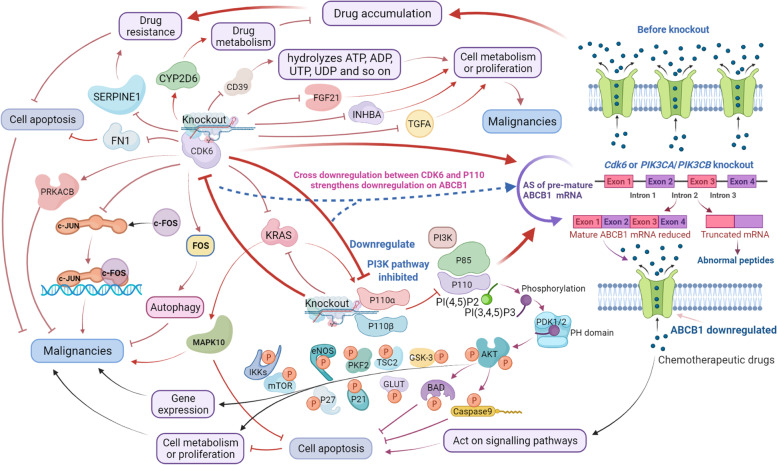
The downregulation of ABCB1 induced by CDK6 deficiency in KB-C2-k.o.cdk6 attracted our attention. In order to uncover the pathway for CDK6 to downregulate ABCB1 expression and reverse MDR, we analyzed differential gene expression based on the transcriptome sequencing database of the KB-C2-k.o.cdk6 cell population. Among the 8974 genes with significant variance in gene expression (p-adjust < 0.001, differential folds > 1.3), the Top10 KEGG pathways involved in the differential gene expression caused by knockout of CDK6 are intimately related to advanced glycation end product (AGE)-AGE receptor (AGE-RAGE) signaling, transforming growth factor-β signaling, endocrine resistance (acquired drug resistance to endocrine therapy of cancers), tumor necrosis factor signaling, et al., among which CDK6 is one of the top10 factors (Fig. 3A). Among the proteins most correlated with endocrine resistance, neurogenic locus notch homolog protein 3 (NOTCH3), transcription factor AP-1 (JUN), and mitogen-activated protein kinase 10 (MAPK10) were downregulated, whereas phosphatidylinositol 3-kinase regulatory subunit gamma (PIK3R3), G-protein coupled estrogen receptor 1 (GPER1), cyclin-dependent kinase inhibitor 2C (CDKN2C), cytochrome P450 2D6 (CYP2D6), cAMP-dependent protein kinase catalytic subunit beta (PRKACB), phosphatidylinositol 3-kinase regulatory subunit beta (PIK3R2) and notch 2 N-terminal like B (NOTCH2NLB) were upregulated. Interactions between 1) MAPK10 and JUN, 2) PIK3R2 and PIK3R3 were revealed (Fig. S1). We then selected the “drug resistance” associated genes from KEGG category. Network analysis showed that they belong to one or more groups associated with different types of resistance or response to drugs or stimuli (Fig. 3B). The differential expression of these genes in KB-C2 and KB-C2-k.o.cdk6 are shown in Fig. 3C. Protein-protein interaction (PPI) analysis revealed: 1) the signaling pathways involving CDK4, FOS, PRKACB, brain V-type proton ATPase subunit B2 (ATP6V1B2), mitochondrial ATP synthase subunit e (ATP5ME) and mitochondrial ATP synthase-coupling factor 6 (ATP5PF), and 2) PI3K/HRAS signaling pathways. However, no regulation between these genes and ABCB1 expression was retrieved, either using metascape or STRING platform (Fig. 3D, Fig. S2). Nevertheless, the experimental data showed that a series of endocrine resistance-related signaling factors were upregulated or downregulated in the cdk6-deficient cells. Among them, FOS-promoting cell autophagy [38] was upregulated, whereas PI3K (PIK3CA/PIK3CB), which is a key signaling factor in cancer and was found to regulate ABCB1-mediated MDR in our recent study [2], was downregulated accompanying the remarkable downregulation of ABCB1 (Fig. 4A, B and C).
Fig. 3.
Category of the differential expression of genes and networks of the selected genes including the drug-resistant genes in KB-C2 and KB-C2-k.o.cdk6 cells. A Top10 KEGG pathways involved in the differential gene expression caused by knockout of cdk6 in KB-C2. B Network of enriched genes. The items are colored by cluster ID, where nodes that share the same cluster ID are typically close to each other. C Differential degree of the drug-resistant and -selected genes in KB-C2-k.o.cdk6 cells. D Protein-protein interaction network of the proteins that are most correlated with acquired drug resistance to endocrine therapy of cancers
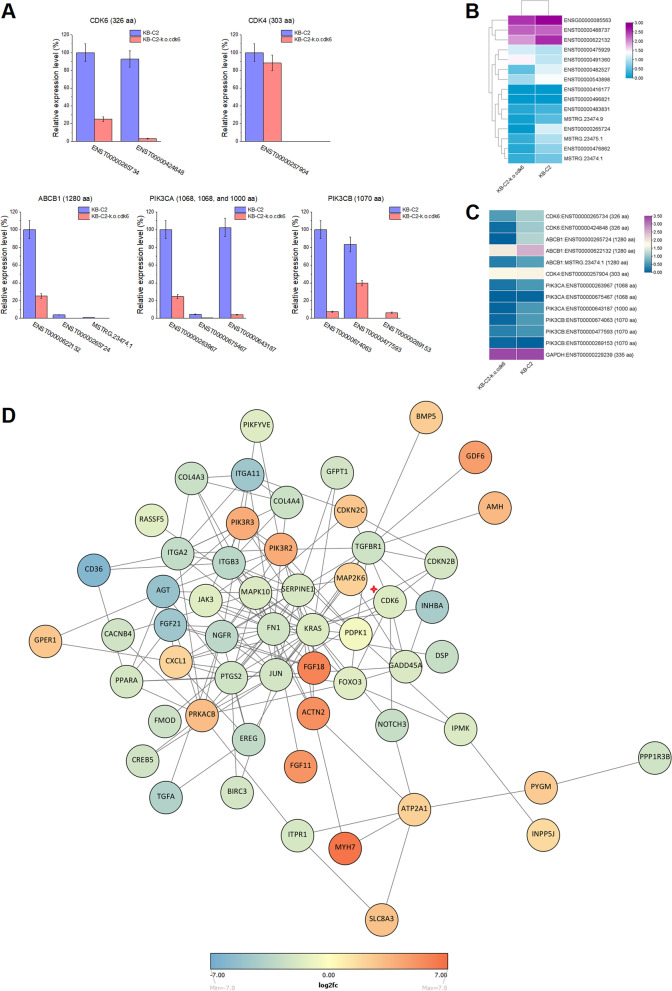
Fig. 4.
Cross-downregulation of CDK6-PI3K axis in the cdk6-deficient KB-C2 cells reduced the expression of ABCB1 by inducing responsive alternative splicing (AS) in the pre-mRNA of ABCB1. A Relative expression of the functional proteins including CDK6, CDK4, ABCB1, PIK3CA and PIK3CB in KB-C2 and the cdk6-deficient KB-C2 cell populations. Major transcripts according to each intact protein were calculated in the cells expressing identical level of GAPDH. B Heatmap showed that through AS of pre-mRNA in response to cdk6 deletion, the level of the transcripts expressing functional ABCB1 protein was significantly downregulated whereas some transcripts expressing truncated inactive ABCB1 peptides (e.g., ENST00000488737) were upregulated in the KB-C2 cells with deleted cdk6 gene. C Differential expression heatmap showing downregulation of PI3K 110α and 110β (encoded by PIK3CA and PIK3CB genes, respectively) in the KB-C2-k.o.cdk6 population as compared with the non-gene deficient KB-C2 cells. D Protein-protein interaction network showing the pathways linking the differently expressed proteins in the KB-C2 cells with deficiency of cdk6. This network reflects the factors that have most intimate relationships with CDK6, and the pathways that are regulated by deletion of cdk6 and belong to different biological functions. Yellow represents the protein with no significant difference in expression. From green to blue, the downregulation of proteins is gradually obvious. From orange to red, the up regulation of proteins is gradually obvious
Cross-downregulation between CDK6 and PI3K 110α/110β suppresses ABCB1 expression in cdk6 deficient cancer cells
Our previous study revealed that P110α and P110β could be potential targets that regulate ABCB1-mediated MDR in cancers [2]. It is of great interest whether cdk6 gene downregulation may cause co-downregulation of PI3K 110α and 110β expression. In so doing, both P110α and P110β expression levels are significantly downregulated with the knockout of cdk6 gene (Fig. 4A and C, Table S1). Therefore, downregulation of ABCB1 expression and reversal of ABCB1-mediated MDR in KB-C2-k.o.cdk6 could possibly, at least partially be induced by the downregulation of P110α and P110β resulted from knocking out the cdk6 gene.
Furthermore, knockout of cdk6, PIK3CA or PIK3CB in KB-C2 indicated that CDK6 and P110α/P110β could positively regulate each other (Tables S1, S2). This reverse regulation of P110α/P110β on CDK6 was also found in H460/MX80 cells via knockout of PIK3CA or PIK3CB genes (Table S2), and further strengthened the possibility that cross regulation exists between CDK6 and P110α/P110β, which co-interact and synergize to regulate ABCB1 expression and ABCB1-mediated MDR in KB-C2 and H460/MX80 cells (Fig. S2).
CDK6, CDK4 and PI3K 110α/110β regulate ABCB1 expression by inducing AS of pre-mRNA of ABCB1 gene
By using the TPM (transcripts per million mapped reads) level of GAPDH encoding transcript (ENST00000229239) as the standard, the levels of transcripts encoding native functional CDK6 (326 aa), i.e. ENST00000265734 and ENST00000424848, were dramatically decreased in KB-C2-k.o.cdk6 cell populations, and 25 and 3%, respectively, remained as compared with those detected in non-gene-deficient KB-C2 cells (Fig. 4A,C Table S1). The ABCB1 transcripts ENST00000622132 and ENST00000265724 encoding functional ABCB1 (1280 aa) protein decreased to 25 and 0%, respectively, in KB-C2-k.o.cdk6 as compared with KB-C2 MDR cells (Fig. 4B, Table S1).
To study the mechanisms of the altered ABCB1 expression induced by CDK6 or CDK4, we analyzed the AS of the pre-mature mRNA of ABCB1 gene, which expresses protein ABCB1. The results demonstrated that in either KB-C2 cell population with cdk6 or cdk4 deficiency, a high frequency of skipped exon (SE) splicing was detected in the ABCB1 pre-mRNA (Table 1). Among the 14 SE events in ABCB1 pre-mRNA of KB-C2-k.o.cdk6 cells, 12 of them were novel chromosomal AS events. Thirteen SE were found in KB-C2-k.o.cdk4 cells, where 10 were novel chromosomal AS. The high frequency of AS events in ABCB1 pre-mRNA in response to cdk6 or cdk4 deficiency generated altered expression level of the ABCB1 protein, further demonstrating that CDK6 and CDK4 may regulate ABCB1 expression by generating responsive AS in ABCB1 pre-mRNA.
Table 1.
Skipped exon (SE) evidence of ABCB1 transcripts detected in transcriptomes from KB-C2-k.o.cdk6, KB-C2-k.o.cdk4, KB-C2-110α, KB-C2-110β as compared with KB-C2 transcriptome
| (I) KB-C2-k.o.cdk6 vs KB-C2 | ||||||||||||
| AS ID | Gene ID | Gene name | Novel AS | Chr | Diff significant | P Value Junction Count Only | Exon Start (0 base) | Exon End | Upstream ES | Upstream EE | Downstream ES | Downstream EE |
| SE_46658 | ENSG00000085563 | ABCB1 | yes | 7 | no | 1 | 87516508 | 87516665 | 87515227 | 87515428 | 87519325 | 87519466 |
| SE_46659 | ENSG00000085563 | ABCB1 | yes | 7 | no | 1 | 87536457 | 87536541 | 87531293 | 87531497 | 87539267 | 87539345 |
| SE_46657 | ENSG00000085563 | ABCB1 | yes | 7 | no | 1 | 87505896 | 87506043 | 87502690 | 87504449 | 87509274 | 87509481 |
| SE_46669 | ENSG00000085563 | ABCB1 | no | 7 | no | 0.9038720596 | 87602959 | 87603140 | 87600812 | 87601078 | 87713160 | 87713295 |
| SE_46668 | ENSG00000085563 | ABCB1 | yes | 7 | yes | 7.74577644824E-006 | 87601966 | 87603140 | 87600754 | 87601078 | 87605645 | 87605775 |
| SE_46663 | ENSG00000085563 | ABCB1 | yes | 7 | no | 1 | 87550724 | 87550838 | 87550467 | 87550578 | 87553760 | 87553932 |
| SE_46662 | ENSG00000085563 | ABCB1 | yes | 7 | no | 1 | 87550170 | 87550296 | 87549850 | 87550054 | 87550467 | 87550578 |
| SE_46661 | ENSG00000085563 | ABCB1 | yes | 7 | no | 1 | 87549347 | 87549518 | 87545862 | 87546024 | 87549850 | 87550054 |
| SE_46660 | ENSG00000085563 | ABCB1 | yes | 7 | no | 1 | 87541356 | 87541464 | 87539267 | 87539345 | 87544128 | 87544275 |
| SE_46667 | ENSG00000085563 | ABCB1 | yes | 7 | no | 1 | 87595765 | 87595814 | 87585511 | 87585680 | 87600116 | 87600190 |
| SE_46666 | ENSG00000085563 | ABCB1 | yes | 7 | no | 1 | 87595765 | 87595814 | 87505896 | 87506043 | 87600116 | 87600190 |
| SE_46665 | ENSG00000085563 | ABCB1 | yes | 7 | no | 0.0682123258 | 87566784 | 87566976 | 87566069 | 87566241 | 87570171 | 87570223 |
| SE_46664 | ENSG00000085563 | ABCB1 | yes | 7 | no | 0.4481719285 | 87566069 | 87566241 | 87561262 | 87561387 | 87570171 | 87570223 |
| SE_46670 | ENSG00000085563 | ABCB1 | no | 7 | no | 0.7879487815 | 87605645 | 87605775 | 87600754 | 87601078 | 87713160 | 87713294 |
| (II) KB-C2-k.o.cdk4 vs KB-C2 | ||||||||||||
| AS ID | Gene ID | Gene name | Novel AS | Chr | Diff significant | P Value Junction Count Only | Exon Start (0 base) | Exon End | Upstream ES | Upstream EE | Downstream ES | Downstream EE |
| SE_47404 | ENSG00000085563 | ABCB1 | yes | 7 | no | 0.1867192198 | 87566784 | 87566976 | 87566069 | 87566241 | 87570171 | 87570223 |
| SE_47405 | ENSG00000085563 | ABCB1 | yes | 7 | no | 1 | 87595765 | 87595814 | 87585511 | 87585680 | 87600116 | 87600190 |
| SE_47406 | ENSG00000085563 | ABCB1 | yes | 7 | no | 0.4757658641 | 87601966 | 87603140 | 87600754 | 87601078 | 87605645 | 87605775 |
| SE_47407 | ENSG00000085563 | ABCB1 | no | 7 | no | 0.1348013017 | 87602959 | 87603140 | 87600812 | 87601078 | 87713160 | 87713295 |
| SE_47400 | ENSG00000085563 | ABCB1 | yes | 7 | no | 1 | 87549347 | 87549518 | 87545862 | 87546024 | 87549850 | 87550054 |
| SE_47401 | ENSG00000085563 | ABCB1 | yes | 7 | no | 1 | 87550170 | 87550296 | 87549850 | 87550054 | 87550467 | 87550578 |
| SE_47402 | ENSG00000085563 | ABCB1 | yes | 7 | no | 1 | 87550724 | 87550838 | 87550467 | 87550578 | 87553760 | 87553932 |
| SE_47403 | ENSG00000085563 | ABCB1 | yes | 7 | no | 0.3242107276 | 87566069 | 87566241 | 87561262 | 87561387 | 87570171 | 87570223 |
| SE_47408 | ENSG00000085563 | ABCB1 | no | 7 | no | 0.0695066227 | 87605645 | 87605775 | 87600754 | 87601078 | 87713160 | 87713294 |
| SE_47409 | ENSG00000085563 | ABCB1 | no | 7 | yes | 2.4192194914E-009 | 87605645 | 87605775 | 87601966 | 87603140 | 87713160 | 87713294 |
| SE_47397 | ENSG00000085563 | ABCB1 | yes | 7 | no | 1 | 87519325 | 87519466 | 87516508 | 87516665 | 87520775 | 87520876 |
| SE_47398 | ENSG00000085563 | ABCB1 | yes | 7 | no | 1 | 87536457 | 87536541 | 87531293 | 87531497 | 87539267 | 87539345 |
| SE_47399 | ENSG00000085563 | ABCB1 | yes | 7 | no | 1 | 87541356 | 87541464 | 87539267 | 87539345 | 87544128 | 87544275 |
| (III) KB-C2-k.o.110α vs KB-C2 | ||||||||||||
| AS ID | Gene ID | Gene name | Novel AS | Chr | Diff significant | P Value Junction Count Only | Exon Start (0 base) | Exon End | Upstream ES | Upstream EE | Downstream ES | Downstream EE |
| SE_50129 | ENSG00000085563 | ABCB1 | no | 7 | no | 0.1120059639 | 87605645 | 87605775 | 87601966 | 87603140 | 87713160 | 87715224 |
| SE_50128 | ENSG00000085563 | ABCB1 | no | 7 | yes | 2.83848985472E-006 | 87605645 | 87605775 | 87600754 | 87601078 | 87713160 | 87715224 |
| SE_50127 | ENSG00000085563 | ABCB1 | no | 7 | yes | 0 | 87602959 | 87603140 | 87600754 | 87601078 | 87713160 | 87713295 |
| SE_50125 | ENSG00000085563 | ABCB1 | no | 7 | no | 0.0641212769 | 87601272 | 87601371 | 87600754 | 87601078 | 87602959 | 87603140 |
| SE_50123 | ENSG00000085563 | ABCB1 | yes | 7 | no | 1 | 87595765 | 87595814 | 87585511 | 87585680 | 87600116 | 87600190 |
| SE_50122 | ENSG00000085563 | ABCB1 | yes | 7 | no | 1 | 87570171 | 87570223 | 87561262 | 87561387 | 87585511 | 87585680 |
| SE_50121 | ENSG00000085563 | ABCB1 | yes | 7 | no | 0.0842138339 | 87566784 | 87566976 | 87566069 | 87566241 | 87570171 | 87570223 |
| SE_50112 | ENSG00000085563 | ABCB1 | yes | 7 | no | 1 | 87519325 | 87519466 | 87516508 | 87516665 | 87520775 | 87520876 |
| SE_50116 | ENSG00000085563 | ABCB1 | yes | 7 | no | 1 | 87550170 | 87550296 | 87549850 | 87550054 | 87550467 | 87550578 |
| SE_50117 | ENSG00000085563 | ABCB1 | yes | 7 | no | 1 | 87550724 | 87550838 | 87550467 | 87550578 | 87553760 | 87553932 |
| SE_50114 | ENSG00000085563 | ABCB1 | yes | 7 | no | 0.5111213437 | 87541356 | 87541464 | 87539267 | 87539345 | 87544128 | 87544275 |
| SE_50126 | ENSG00000085563 | ABCB1 | yes | 7 | no | 0.2905363226 | 87601966 | 87603140 | 87600754 | 87601078 | 87605645 | 87605775 |
| SE_50124 | ENSG00000085563 | ABCB1 | yes | 7 | no | 0.2672547758 | 87601272 | 87601371 | 87600754 | 87601078 | 87601966 | 87603140 |
| SE_50120 | ENSG00000085563 | ABCB1 | yes | 7 | no | 0.5376920457 | 87566784 | 87566976 | 87561262 | 87561387 | 87570171 | 87570223 |
| SE_50113 | ENSG00000085563 | ABCB1 | yes | 7 | no | 1 | 87536457 | 87536541 | 87531293 | 87531497 | 87539267 | 87539345 |
| SE_50115 | ENSG00000085563 | ABCB1 | yes | 7 | no | 1 | 87549347 | 87549518 | 87545862 | 87546024 | 87549850 | 87550054 |
| SE_50118 | ENSG00000085563 | ABCB1 | yes | 7 | no | 1 | 87566069 | 87566241 | 87561262 | 87561387 | 87566784 | 87566976 |
| SE_50119 | ENSG00000085563 | ABCB1 | yes | 7 | no | 0.4481965282 | 87566069 | 87566241 | 87561262 | 87561387 | 87570171 | 87570223 |
| (V) KB-C2-k.o.110β vs KB-C2 | ||||||||||||
| AS ID | Gene ID | Gene name | Novel AS | Chr | Diff significant | P Value Junction Count Only | Exon Start (0 base) | Exon End | Upstream ES | Upstream EE | Downstream ES | Downstream EE |
| SE_49198 | ENSG00000085563 | ABCB1 | yes | 7 | no | 0.4057523949 | 87601272 | 87601371 | 87600754 | 87601078 | 87601966 | 87603140 |
| SE_49199 | ENSG00000085563 | ABCB1 | no | 7 | yes | 0.0006903622 | 87601272 | 87601371 | 87600754 | 87601078 | 87602959 | 87603140 |
| SE_49190 | ENSG00000085563 | ABCB1 | yes | 7 | no | 1 | 87550170 | 87550296 | 87549850 | 87550054 | 87550467 | 87550578 |
| SE_49191 | ENSG00000085563 | ABCB1 | yes | 7 | no | 0.365137306 | 87550467 | 87550578 | 87549850 | 87550054 | 87553760 | 87553932 |
| SE_49192 | ENSG00000085563 | ABCB1 | yes | 7 | no | 1 | 87550724 | 87550838 | 87550467 | 87550578 | 87553760 | 87553932 |
| SE_49193 | ENSG00000085563 | ABCB1 | yes | 7 | no | 1 | 87566069 | 87566241 | 87561262 | 87561387 | 87566784 | 87566976 |
| SE_49194 | ENSG00000085563 | ABCB1 | yes | 7 | no | 0.448132556 | 87566069 | 87566241 | 87561262 | 87561387 | 87570171 | 87570223 |
| SE_49195 | ENSG00000085563 | ABCB1 | yes | 7 | no | 0.5487589032 | 87566784 | 87566976 | 87561262 | 87561387 | 87570171 | 87570223 |
| SE_49196 | ENSG00000085563 | ABCB1 | no | 7 | no | 0.9285527169 | 87566784 | 87566976 | 87566069 | 87566241 | 87570171 | 87570223 |
| SE_49197 | ENSG00000085563 | ABCB1 | yes | 7 | no | 1 | 87595765 | 87595814 | 87585511 | 87585680 | 87600116 | 87600190 |
| SE_49189 | ENSG00000085563 | ABCB1 | yes | 7 | no | 1 | 87549347 | 87549518 | 87545862 | 87546024 | 87549850 | 87550054 |
| SE_49188 | ENSG00000085563 | ABCB1 | yes | 7 | no | 1 | 87541356 | 87541464 | 87539267 | 87539345 | 87544128 | 87544275 |
| SE_49187 | ENSG00000085563 | ABCB1 | yes | 7 | no | 1 | 87536457 | 87536541 | 87531293 | 87531497 | 87539267 | 87539345 |
| SE_49186 | ENSG00000085563 | ABCB1 | yes | 7 | no | 1 | 87516508 | 87516665 | 87515227 | 87515428 | 87519325 | 87519466 |
| SE_49200 | ENSG00000085563 | ABCB1 | no | 7 | yes | 1.1990408666E-014 | 87601272 | 87601371 | 87600754 | 87601078 | 87713160 | 87715224 |
| SE_49201 | ENSG00000085563 | ABCB1 | yes | 7 | yes | 0.000649673 | 87601966 | 87603140 | 87600754 | 87601078 | 87605645 | 87605775 |
| SE_49202 | ENSG00000085563 | ABCB1 | no | 7 | yes | 0 | 87602959 | 87603140 | 87600754 | 87601078 | 87713160 | 87713295 |
| SE_49203 | ENSG00000085563 | ABCB1 | yes | 7 | yes | 0.0067345237 | 87602959 | 87603140 | 87601272 | 87601371 | 87713160 | 87713295 |
| SE_49204 | ENSG00000085563 | ABCB1 | no | 7 | yes | 0 | 87605645 | 87605775 | 87600754 | 87601078 | 87713160 | 87715224 |
| SE_49205 | ENSG00000085563 | ABCB1 | no | 7 | no | 0.0538934718 | 87605645 | 87605775 | 87601966 | 87603140 | 87713160 | 87715224 |
| (I) KB-C2-k.o.cdk6 vs KB-C2 | |||||||||||
| IJC KB-C2 | SJC KB-C2 | IJC KB-C2-k.o.cdk6 | SJC KB-C2-k.o.cdk6 | Inc Form Len | Skip Form Len | Average Inc Level1 (average PSI 1) | Average Inc Level2 (average PSI 2) | Increase Inclusion KB-C2 (ΔPSI>0) | Increase Exclusion KB-C2 (ΔPSI<0) | Increase Inclusion KB-C2-k.o.cdk6 (ΔPSI<0) | Increase Exclusion KB-C2-k.o.cdk6 (ΔPSI>0) |
| 7907 | 9 | 5933 | 0 | 298 | 149 | 0.998 | 1 | no | yes | yes | no |
| 2821 | 1 | 2975 | 0 | 232 | 149 | 0.999 | 1 | no | yes | yes | no |
| 12259 | 1 | 7047 | 0 | 295 | 149 | 1 | 1 | no_difference | no_difference | no_difference | no_difference |
| 11 | 35 | 38 | 129 | 298 | 149 | 0.136 | 0.128 | yes | no | no | yes |
| 0 | 2 | 50 | 1 | 298 | 149 | 0 | 0.962 | no | yes | yes | no |
| 2116 | 2 | 4377 | 3 | 262 | 149 | 0.998 | 0.999 | no | yes | yes | no |
| 2845 | 2 | 4885 | 4 | 274 | 149 | 0.999 | 0.998 | yes | no | no | yes |
| 3787 | 9 | 5847 | 1 | 298 | 149 | 0.995 | 1 | no | yes | yes | no |
| 2626 | 0 | 3200 | 14 | 256 | 149 | 1 | 0.993 | yes | no | no | yes |
| 1170 | 0 | 3565 | 27 | 197 | 149 | 1 | 0.99 | yes | no | no | yes |
| 349 | 1 | 931 | 0 | 197 | 149 | 0.996 | 1 | no | yes | yes | no |
| 1491 | 45 | 4298 | 83 | 298 | 149 | 0.943 | 0.963 | no | yes | yes | no |
| 857 | 0 | 1995 | 14 | 298 | 149 | 1 | 0.986 | yes | no | no | yes |
| 2 | 35 | 4 | 129 | 278 | 149 | 0.03 | 0.016 | yes | no | no | yes |
| (II) KB-C2-k.o.cdk4 vs KB-C2 | |||||||||||
| IJC KB-C2 | SJC KB-C2 | IJC KB-C2-k.o.cdk4 | SJC KB-C2-k.o.cdk4 | Inc Form Len | Skip Form Len | Average Inc Level1 (average PSI 1) | Average Inc Level2 (average PSI 2) | Increase Inclusion KB-C2 (ΔPSI>0) | Increase Exclusion KB-C2 (ΔPSI<0) | Increase Inclusion KB-C2-k.o.cdk4 (ΔPSI<0) | Increase Exclusion KB-C2-k.o.cdk4 (ΔPSI>0) |
| 3892 | 56 | 4300 | 83 | 341 | 149 | 0.968 | 0.958 | yes | no | no | yes |
| 2933 | 7 | 3565 | 27 | 198 | 149 | 0.997 | 0.99 | yes | no | no | yes |
| 54 | 0 | 50 | 1 | 1323 | 149 | 1 | 0.849 | yes | no | no | yes |
| 46 | 101 | 38 | 129 | 330 | 149 | 0.171 | 0.117 | yes | no | no | yes |
| 5497 | 14 | 5847 | 1 | 320 | 149 | 0.995 | 1 | no | yes | yes | no |
| 5076 | 4 | 4885 | 4 | 275 | 149 | 0.999 | 0.998 | yes | no | no | yes |
| 4228 | 0 | 4377 | 3 | 263 | 149 | 1 | 0.999 | yes | no | no | yes |
| 1914 | 1 | 1995 | 14 | 321 | 149 | 0.999 | 0.985 | yes | no | no | yes |
| 9 | 101 | 4 | 129 | 279 | 149 | 0.045 | 0.016 | yes | no | no | yes |
| 20 | 0 | 3 | 11 | 279 | 149 | 1 | 0.127 | yes | no | no | yes |
| 5188 | 1 | 5459 | 0 | 290 | 149 | 1 | 1 | no_difference | no_difference | no_difference | no_difference |
| 3032 | 4 | 2975 | 0 | 233 | 149 | 0.998 | 1 | no | yes | yes | no |
| 3431 | 0 | 3200 | 14 | 257 | 149 | 1 | 0.993 | yes | no | no | yes |
| (III) KB-C2-k.o.110α vs KB-C2 | |||||||||||
| IJC KB-C2 | SJC KB-C2 | IJC KB-C2-k.o.110α | SJC KB-C2-k.o.110α | Inc Form Len | Skip Form Len | Average Inc Level1 (average PSI 1) | Average Inc Level2 (average PSI 2) | Increase Inclusion KB-C2 (ΔPSI>0) | Increase Exclusion KB-C2 (ΔPSI<0) | Increase Inclusion KB-C2-k.o.110α (ΔPSI<0) | Increase Exclusion KB-C2-k.o.110α (ΔPSI>0) |
| 11 | 14 | 3 | 11 | 278 | 149 | 0.296 | 0.128 | yes | no | no | yes |
| 3 | 0 | 4 | 129 | 278 | 149 | 1 | 0.016 | yes | no | no | yes |
| 30 | 0 | 38 | 129 | 298 | 149 | 1 | 0.128 | yes | no | no | yes |
| 38 | 16 | 30 | 27 | 247 | 149 | 0.589 | 0.401 | yes | no | no | yes |
| 4622 | 7 | 3565 | 27 | 197 | 149 | 0.998 | 0.99 | yes | no | no | yes |
| 818 | 1 | 701 | 0 | 200 | 149 | 0.998 | 1 | no | yes | yes | no |
| 5347 | 145 | 4298 | 83 | 298 | 149 | 0.949 | 0.963 | no | yes | yes | no |
| 7533 | 7 | 5459 | 0 | 289 | 149 | 0.998 | 1 | no | yes | yes | no |
| 6308 | 5 | 4885 | 4 | 274 | 149 | 0.999 | 0.998 | yes | no | no | yes |
| 5327 | 2 | 4377 | 3 | 262 | 149 | 0.999 | 0.999 | no_difference | no_difference | no_difference | no_difference |
| 4489 | 32 | 3200 | 14 | 256 | 149 | 0.988 | 0.993 | no | yes | yes | no |
| 58 | 0 | 50 | 1 | 298 | 149 | 1 | 0.962 | yes | no | no | yes |
| 66 | 50 | 47 | 50 | 247 | 149 | 0.443 | 0.362 | yes | no | no | yes |
| 2856 | 2 | 2099 | 14 | 298 | 149 | 0.999 | 0.987 | yes | no | no | yes |
| 4051 | 26 | 2975 | 0 | 232 | 149 | 0.99 | 1 | no | yes | yes | no |
| 8098 | 9 | 5847 | 1 | 298 | 149 | 0.998 | 1 | no | yes | yes | no |
| 4749 | 1 | 4111 | 0 | 298 | 149 | 1 | 1 | no_difference | no_difference | no_difference | no_difference |
| 2402 | 2 | 1995 | 14 | 298 | 149 | 0.998 | 0.986 | yes | no | no | yes |
| (V) KB-C2-k.o.110β vs KB-C2 | |||||||||||
| IJC KB-C2 | SJC KB-C2 | IJC KB-C2-k.o.110β | SJC KB-C2-k.o.110β | Inc Form Len | Skip Form Len | Average Inc Level1 (average PSI 1) | Average Inc Level2 (average PSI 2) | Increase Inclusion KB-C2 (ΔPSI>0) | Increase Exclusion KB-C2 (ΔPSI<0) | Increase Inclusion KB-C2-k.o.110β (ΔPSI<0) | Increase Exclusion KB-C2-k.o.110β (ΔPSI>0) |
| 21 | 30 | 47 | 50 | 247 | 149 | 0.297 | 0.362 | no | yes | yes | no |
| 15 | 53 | 30 | 27 | 247 | 149 | 0.146 | 0.401 | no | yes | yes | no |
| 6090 | 5 | 4885 | 4 | 274 | 149 | 0.998 | 0.998 | no_difference | no_difference | no_difference | no_difference |
| 12 | 1 | 5 | 0 | 259 | 149 | 0.873 | 1 | no | yes | yes | no |
| 5428 | 7 | 4377 | 3 | 262 | 149 | 0.998 | 0.999 | no | yes | yes | no |
| 4885 | 1 | 4111 | 0 | 298 | 149 | 1 | 1 | no_difference | no_difference | no_difference | no_difference |
| 2345 | 0 | 1995 | 14 | 298 | 149 | 1 | 0.986 | yes | no | no | yes |
| 2793 | 0 | 2099 | 14 | 298 | 149 | 1 | 0.987 | yes | no | no | yes |
| 5439 | 107 | 4298 | 83 | 298 | 149 | 0.962 | 0.963 | no | yes | yes | no |
| 4078 | 0 | 3565 | 27 | 197 | 149 | 1 | 0.99 | yes | no | no | yes |
| 7421 | 5 | 5847 | 1 | 298 | 149 | 0.999 | 1 | no | yes | yes | no |
| 4110 | 0 | 3200 | 14 | 256 | 149 | 1 | 0.993 | yes | no | no | yes |
| 3805 | 9 | 2975 | 0 | 232 | 149 | 0.996 | 1 | no | yes | yes | no |
| 8034 | 1 | 5933 | 0 | 298 | 149 | 1 | 1 | no_difference | no_difference | no_difference | no_difference |
| 19 | 0 | 28 | 129 | 247 | 149 | 1 | 0.116 | yes | no | no | yes |
| 53 | 12 | 50 | 1 | 298 | 149 | 0.688 | 0.962 | no | yes | yes | no |
| 109 | 0 | 38 | 129 | 298 | 149 | 1 | 0.128 | yes | no | no | yes |
| 58 | 6 | 15 | 0 | 298 | 149 | 0.829 | 1 | no | yes | yes | no |
| 33 | 0 | 4 | 129 | 278 | 149 | 1 | 0.016 | yes | no | no | yes |
| 44 | 56 | 3 | 11 | 278 | 149 | 0.296 | 0.128 | yes | no | no | yes |
The knockout of either PIK3CA (encoding P110α) or PIK3CB (encoding P110β) in KB-C2 cells also resulted in a number of novel AS events in ABCB1 pre-mRNA and downregulation of ABCB1 protein expression (Table 1). Interestingly, 10 common SE positions in ABCB1 were found in the four types of gene-deficient cell populations, i.e., KB-C2 cells deficient in cdk6, cdk4, PIK3CA and PIK3CB (Table 1). They correspond to 87,550,170–87,550,296, 87,536,457–87,536,541, 87,541,356–87,541,464, 87,549,347–87,549,518, 87,550,724–87,550,838, 87,566,069–87,566,241, 87,566,784–87,566,976, 87,595,765–87,595,814, 87,601,966–87,603,140, 87,605,645–87,605,775 on Chromosome 7 (Table 1). These positions could be responsible for the high frequency of AS in ABCB1 pre-mRNA, leading to changes in ABCB1 expression in these gene-deficient cell populations.
Profile of the protein-protein interaction network linking the most differently expressed factors in KB-C2 cells deficient in cdk6
Transcriptome sequencing and quantification revealed that the knockout of cdk6 gene (indicated by a red star in Fig. 4D) induced downregulation of KRAS, CDKN2B, NOTCH3, GADD45A, JUN, FOXO3, TGFBR1, and upregulation of CDKN2C, further induced downregulation of a series of factors, such as PDPK1, SERPINE1, PTGS2, MAPK10, RASSF5, PTGS2 and NGFR, and upregulation of PIK3R2, PIK3R3, and PRKACB (Fig. 4D). The majority of the signaling factors were downregulated. CD36, ITGA11, AGT, FGF21, INHBA and TGFA were the most significantly downregulated factors. FGF18, ACTN2, FGF11, GDF6, PIK3R3, PIK3R2 and MYH7 were the most significantly upregulated ones (Fig. 4D). Among the downregulated factors, KRAS, JUN, MAPK10, FN1 and SERPINE1 are in a relatively central position, and regulates multiple pathways (Fig. 4D). Among the upregulated factors, ATP2A2, PRKACB, PIK3R2, PIK3R3, and CXCL1 regulate multiple pathways (Fig. 4D).
Among the significantly downregulated proteins, KRAS is a GTPase and a proto-oncogene product relaying early signals from outside the cell to the cell nucleus [39]. It is one of the effectors downstream of EGFR activation and triggers intracellular signaling cascades such as MAPK and PI3K [40]. It is interesting that KRAS was significantly downregulated in either CDK6 or P100α/P110β deficient cell populations. The JUN product, proto-oncoprotein c-JUN, in combination with c-Fos, forms oncogenic transcription factor that interacts with specific target DNA sequence to regulate gene expression and is involved in both translocations and deletions in human malignancies [41]. Mitogen-activated protein kinase 10 (MAPK10), also known as c-Jun N-terminal kinase 3 (JNK3) is a c-Jun N-terminal kinases (JNKs) that may prevent cell apoptosis. MAPK10/JNK3 signaling may induce carcinogenesis [42]. Knocking down MAPK10 suppressed ovarian cancer cell growth and migration [43]. Fibronectin 1 (FN1) is a glycoprotein of the extracellular matrix that binds to integrins (a type of membrane-spanning receptor proteins) [44]. It can suppress apoptosis and promoted viability, invasion, and migration in cancers [45]. SERPINE1 gene in human encodes serpin E1, also known as endothelial plasminogen activator inhibitor or plasminogen activator inhibitor-1 (PAI-1), which has significant functions in the cancer occurrence, relapse and multidrug resistance [46]. Therefore, the knockout of cdk6 gene inducing remarkable downregulation of these genes (Fig. 5) may contribute to the inhibition on the survivability or proliferation of the KB-C2 cells.
Fig. 5.
Regulation of CDK6-PI3K axis on ABCB1-mediated drug resistance. Knockout of cdk6 gene in KB-C2 with CRISPR/Cas9 technique induced attenuation of ABCB1-mediated drug resistance. It promoted AS in the pre-mRNA expressing ABCB1, generating differential expression of the transcripts encoding functional ABCB1 protein or truncated abnormal peptides, further decreased the level of functional ABCB1 transporters. With the deficiency of cdk6 in KB-C2 cells, PI3K 110α and 110β expression were downregulated, which induced AS of ABCB1 pre-mRNA and led to inhibition of ABCB1 expression and reversal of ABCB1-mediated MDR, inhibiting cell division and promoting cell apoptosis. A series of signaling pathways critical for malignancies were downregulated in the KB-C2-k.o.cdk6 cell population
Among the significantly upregulated proteins, ATP2A2 as one of the P-type pumps important for higher order physiological processes, is an isoform of the SERCA Ca2+-ATPases that catalyze the hydrolysis of ATP, maintain nanomolar cytoplasmic Ca2+ levels, and provide high levels of Ca2+ in the endoplasmic reticulum, Golgi, and secretory vesicles which store Ca2+ for a wide range of signaling functions [47, 48]. cAMP-dependent protein kinase catalytic subunit beta (PRKACB) is a member of the serine/threonine protein kinase family and is a catalytic subunit of PKA. By activating the protein kinase A (PKA), which transduces the signal through phosphorylation of variant target proteins, it functions as a signaling molecule important for a variety of cellular activities [49]. As reported, down-regulation of PRKACB is related to the occurrence of various human malignancies [50]. the PI3K regulatory subunits PIK3R2/p85beta and PIK3R3/p55gamma, are frequently overexpressed in various cancers [51, 52]. The chemokine (C-X-C motif) ligand 1 (CXCL1) is a small peptide belonging to the CXC chemokine family. It promotes arteriogenesis through enhanced monocyte recruitment into the peri-collateral space and has relationship with tumor progression [53, 54]. For these significantly upregulated genes that have central regulation function and are involved in multiple signaling pathways (Fig. 4D), ATP2A2 and PRKACB upregulation may contribute to inhibiting the viability of KB-C2 cells. It is also the first time to unveil the significant upregulation of some malignancy related genes including PIK3R2, PIK3R3 and CXCL1. These results indicated that responsive upregulation or downregulation of a number of genes and signaling pathways occurred that affected multiple cell functions after cdk6 knockout. The mostly changed signaling pathways downregulating ABCB1 expression and inhibiting cancer cell proliferation were concluded in Fig. 5.
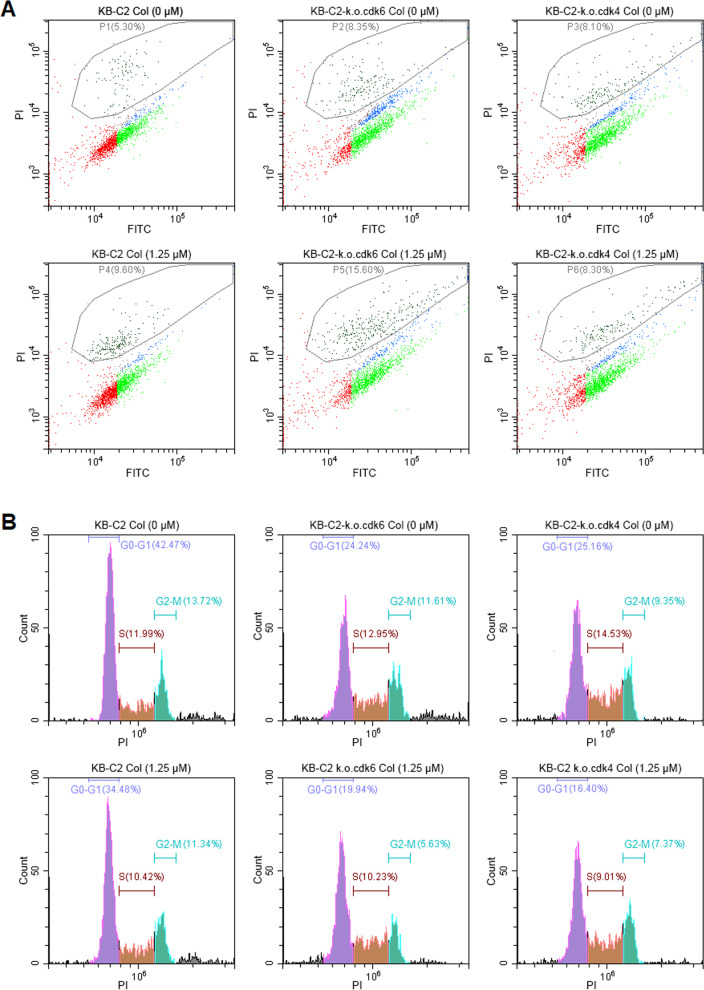
Knockout of cdk6 or cdk4 gene promotes apoptosis, but only cdk6 deficiency reverses MDR in KB-C2 cells
In the cdk4- or cdk6-deficient KB-C2 cells cultured with or without colchicine, the proportion of non-apoptotic cells was half declined compared with the original KB-C2 cells (Fig. 6A). The deletion of the cdk6 gene induced transition of more cells from slightly apoptotic status to heavily apoptotic status (Fig. 6).
Fig. 6.
Flow cytometer analysis of cell apoptosis and cell cycle in KB-C2 cells with the deletion of cdk6 or cdk4 gene.A Apoptosis of KB-C2 cells with deletion of cdk6 or cdk4 gene. The KB-C2-k.o.cdk6 and KB-C2-k.o.cdk4 cells were either incubated with a vehicle (media without colchicine) or colchicine (Col) for 24 h. The fluorescent intensity of the cells at the various heights on the axis are presented as spot groups showing the percentage of viable to severely apoptotic cell populations. The populations comprising heavily apoptotic (HA) cells (P1-P6), moderately apoptotic (MA) cells, slightly apoptotic (SA) cells and non-apoptotic (NA) cells were indicated by dots in black, blue, green, and red, sequentially. Col: colchicine. The fluorescence of PI and FITC was determined through PC7-A and FITC-A tracks, respectively. B Alteration of cell cycle in the KB-C2 cells with the deletion of cdk6 or cdk4 gene. The cells were co-incubated with the vehicle or colchicine (Col) for 24 h. The fluorescence of PI was determined through PE-A track
The culture of KB-C2 cells with colchicine (1.25 μM) only produced minor increase of apoptotic cells because KB-C2 cancer cells are highly resistant to colchicine (Fig. 6). However, cdk6 knockout remarkably increased the drug sensitivity and induced significantly promoted transition from moderately apoptotic cells to heavily apoptotic cells (15.6%) in KB-C2-k.o.cdk6 cells incubated with colchicine compared with control cells incubated without colchicine (8.35%) (Fig. 6). Slightly increase of non-apoptotic cells and decrease of moderately apoptotic cells was detected in the drug-treated KB-C2-k.o.cdk4 cell populations (cdk4 deleted KB-C2) compared with control cells (Fig. 6).
The deletion of cdk6 or cdk4 accelerates cell senescence in the G1 phase, whereas cdk6 deletion remarkably decreased the drug resistance of KB-C2 cells to colchicine
Deletion of either cdk4 or cdk6 accelerated senescence in G1 phase and promoted apoptosis of KB-C2 cells (Fig. 6B, Table S3). However, the deficiency of cdk6 but not cdk4, caused significant inhibition on cell division in G2-M phase in the presence of colchicine compared with non-gene-deficient KB-C2 cells. This further demonstrated that the knockout of ckd6 gene but not cdk4 reversed the MDR of KB-C2 via a mechanism of inhibiting cell division. As reported, colchicine binds to microtubules that form the mitotic spindle during cell division, thereby leads to mitotic arrest and ultimately cause cell death [55]. Downregulation of ABCB1 by the deficiency of CDK6 may reduce the efflux of colchicine which results in the increase of colchicine accumulation within the cells and enhanced inhibition on cell division.
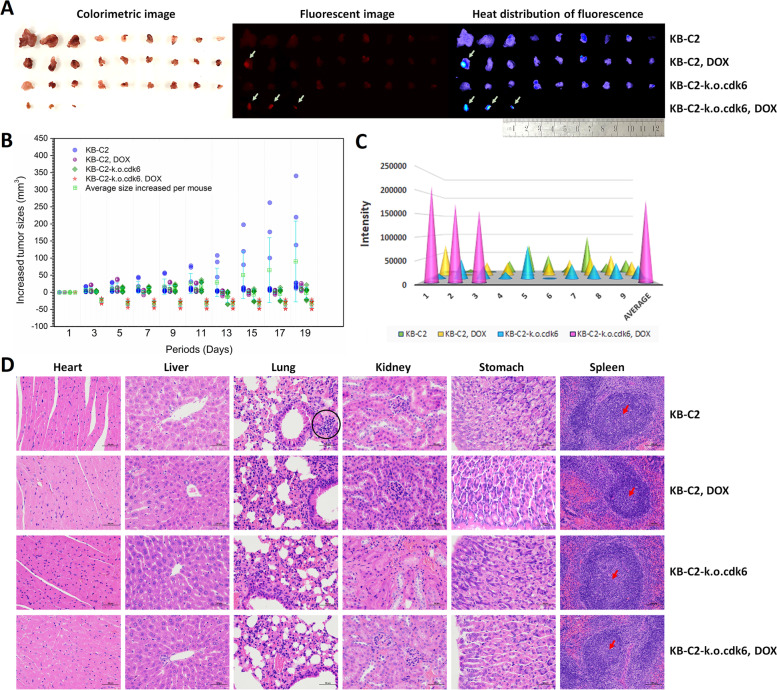
Loss of gene cdk6 remarkably increased the sensitivity of KB-C2 tumors to DOX by increasing DOX accumulation inside the tumor cells
DOX remarkably inhibited the cdk6 deficient KB-C2, i.e., KB-C2-k.o.cdk6 tumors. Among the 9 KB-C2-k.o.cdk6 tumors, 6 were completely eliminated, and only 3 with very small sizes were detected (Fig. 7A and B) by the 19th day after DOX treatment. Tumor enlargement was most rapid in DOX-untreated KB-C2 group, followed by DOX-treated KB-C2 and DOX-untreated KB-C2-k.o.cdk6 groups. This result indicated that loss of gene cdk6 remarkably increased the sensitivity of KB-C2 tumors to DOX. Meanwhile, cdk6 deficiency can inhibit tumor growth to some extent because this gene is important for cancer cell proliferation.
Fig. 7.
In vivo study of the function of gene cdk6 to regulate drug-resistance of KB-C2 tumors. A Image of the tumors from the tumor-bearing mice showing the tumor morphology and the fluorescent DOX (depicted with yellow-green arrows) accumulated in the cdk6-present or cdk6-deficient tumors. The mice treated with saline were set as the blank control group. The tumors were sampled by the 19th day of DOX treatment. B Increased tumor sizes after the mice received chemotherapy using DOX. The mice treated with saline were set as the blank control group. C Comparison of the relative fluorescent intensity showing the difference of DOX accumulation in the cdk6-present or cdk6-deficient tumors. D Pathology of main organs of the mice bearing KB-C2 or KB-C2-k.o.cdk6 tumors. The organs were sampled by the 19th day after DOX treatment. The circle in black depicts focal infiltration of peri-bronchial inflammatory cells. The arrows in red depicts white pulp germinal center expansion, which occurred frequently in the tumor-bearing mice untreated with DOX, but only occasionally detected in the DOX-treated mice bearing KB-C2-k.o.cdk6
To find out the mechanisms of the enhanced inhibition in KB-C2-k.o.cdk6 tumors, the drug sensitivity to DOX was compared between the cdk6-present and cdk6-deficient tumors. All remaining KB-C2-k.o.cdk6 tumors showed strong fluorescence of DOX, while only a small part of the KB-C2 tumors had detectable DOX accumulation (Fig. 7C). This result demonstrated that cdk6 deficiency greatly reversed the drug resistance caused by ABCB1, of which DOX is its substrate, as more amount of DOX was concentrated within the tumor cells where ABCB1 was downregulated by cdk6 deficiency.
With the inhibition of tumor growth, pathological changes including focal infiltration of peri-bronchial inflammatory cells and expansion of the germinal center in spleen white pulp was remarkably reduced in the DOX-treated nude mice bearing KB-C2-k.o.cdk6, as compared with the KB-C2 groups or the DOX-untreated KB-C2-k.o.cdk6 group (Fig. 7D).
Combining knockdown of cdk6 and DOX-chemotherapy inhibited growth and metastasis of KB-C2 tumor in vivo
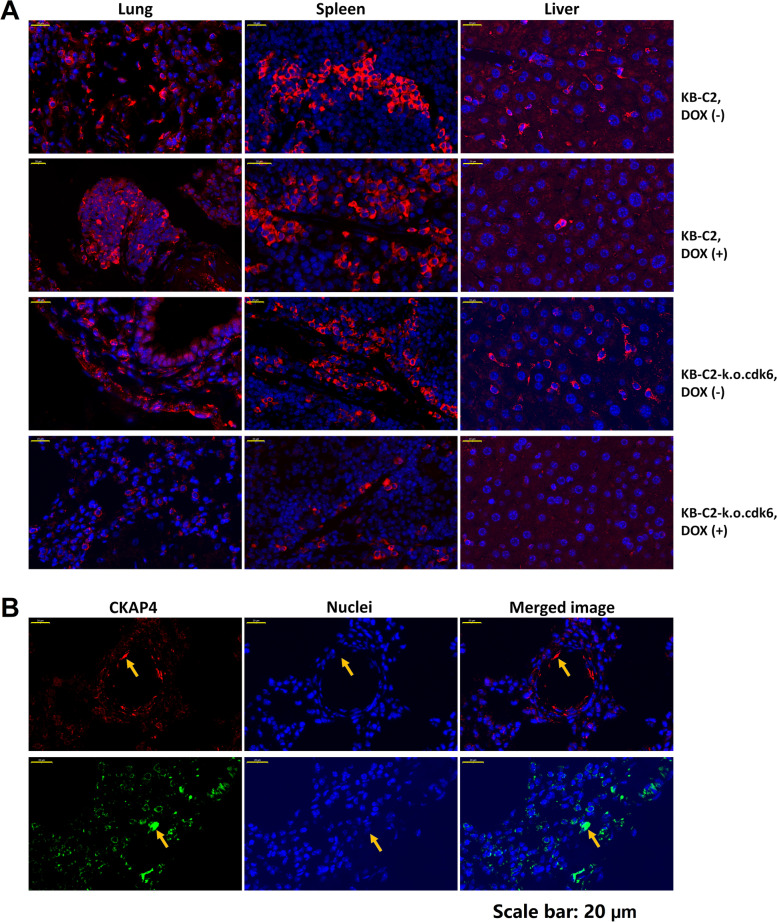
Gene differential expression analysis and immunofluorescence analysis indicated that KB-C2 cells with small cell nuclei and spindle-shaped morphology showed strong CKAP4-positive signal. High levels of tumor cell metastasis were detected in the liver, lung and spleen of KB-C2 tumor-bearing nude mice, and more tumor cells were distributed inside the organs, as compared with KB-C2-k.o.cdk6 tumor- bearing nude mice (Fig. 8). In KB-C2-bearing nude mice treated with DOX, the tumor cells in the organs were only slightly less than those without DOX treatment, and in the lungs, the tumor cells showed obvious inward invasion and growth. In KB-C2-k.o.cdk6-bearing nude mice with DOX treatment, the tumor cells that had invaded in the interior of the organ were the least (Fig. 8), and for the lungs, they were detected only around one outside and no apparent signals were detected inside the lung tissues. These results demonstrated that combining knockdown of cdk6 and DOX chemotherapy inhibited the metastatic ability of MDR KB-C2 cells. Interestingly, KB-C2-k.o.cdk6 cells were eliminated in the lungs of the nude mice after they received DOX by injection.
Fig. 8.
Migration of cdk6 intact and deficient KB-C2 cells to lungs, spleens and livers in the nude mice. A The results of immunofluorescence labeling of CKAP4 showed that the cdk6-deleted cancer cells in the nude mice injected with DOX were the least in number of metastasis spots. DOX had little effect on the metastasis of cancer cells without cdk6 deletion, and had a significant effect on the metastasis of cdk6-deleted cancer cells. All nude mice without DOX injection had serious cancer cell metastasis. B KB-C2-k.o.cdk6 cells were eliminated from the lungs in nude mice after treatment with DOX. The arrows in orange depict the distorted dead KB-C2-k.o.cdk6 cells that had expressed high level of CKAP4. Due to the damage of the nuclei, DAPI fluorescence was absent in these cells. To testify this phenomenon, red and green fluorescence, respective of Cy3 and FITC labeled secondary antibodies, were used to determine antigen CKAP4-abundant KB-C2-k.o.cdk6 cells
Discussion
MDR of cancers can induce failure of chemotherapy. As one major reason of MDR in cancers, ABC transporters are highly expressed in various MDR cancer cell lines [56, 57]. Today, a number of ABC transporter inhibitors have been explored to attenuate the activity of the ABC transporters to pump out chemotherapeutic drugs [2, 58]. However, most of the inhibitors may have several targets, some of which have important function in normal cells [31]. Furthermore, the regulatory mechanisms are greatly unknown; as such risks of side-effects may be increased. Seeking safer targets, often based on tumor-specific proteins that regulate ABC transporters, has become an ideal approach to improve MDR-reversal in cancer therapy. As ABCB1 is a vital ABC transporter for the cancer cells to extrude a variety of anti-cancer drugs [31, 59, 60], we investigated the targets to reverse ABCB1-mediated MDR based on screening of effective reversal agents and gene deletion technology, and revealed a novel targeting pathway. In KB-C2, CDK6 may induce on-off regulation of ABCB1 expression through coordination with PI3K 110α/β and inducing AS in the ABCB1 pre-mRNA. In a parallel study, similar regulation of ABCB1-mediated MDR by CDK6 was observed in H460/MX80-k.o.cdk6, in which cdk6 was knocked out from the NSCLC H460/MX80 MDR cells (Fig. S3). Therefore, CDK6 may be potential and secure targets for reversing ABCB1-mediated MDR, since its overexpression has been shown to stimulate cancer cell proliferation [61].
At present, the project focusing on the function of ribociclib in inhibiting the ABCB1 mediated-MDR in cancers are undergoing in our laboratory. Because the mechanisms for ribociclib to downregulate ABCB1 and inhibit P-gp mediated MDR in cancers could be associated with many aspects, we are currently performing experiments, including real-time PCR of ABCB1 in ribociclib treated cells, or transcriptome sequencing and quantification, in addition to Western blot and immunofluorescence which we have done, to explore the ABCB1 expression at the transcription level. We will also try to explore other potential pathway(s) through the participation of which, ribociclib might downregulate ABCB1 expression. In addition, our unpublished data showed that the structure of ABCB1 could be changed by ribociclib at effective MDR-reversal concentration (e.g., 9 μM) under physiology condition, causing inhibition of the ATPase and drug-efflux activity of ABCB1, indicating that ABCB1 was less stable through interaction with ribociclib. Due to the intricacy of the function of ribociclib within the MDR cancer cells, specific gene deletion was performed to analyze whether CDK6 could be an effective target for overcoming ABCB1-mediated MDR in cancer cells.
We recently realized that the knockout of mutant PI3K 110α or 110β achieved inhibition of ABCB1 expression as well as reversal of MDR mediated by ABCB1 [2]. We further found in this study that downregulation of CDK6 expression through gene knockout downregulated the expression of PI3K P110α and P110β subunits, and vice versa. This cross regulation between CDK6 and P110α/P110β could explain the remarkable decrease in ABCB1 expression and MDR in cdk6 or P110α/P110β deleted KB-C2 cells.
Upon further analysis of the regulatory mechanism of ABCB1 expression by CDK6, CDK4 and PI3K 110 α/β, we surprisingly found that the TPM levels of the ABCB1 transcripts expressing the ABCB1 protein and truncated ABCB1 peptides varied significantly between the gene-deficient KB-C2 cell populations and non-gene-deficient KB-C2 cells. In addition, we found that a high frequency of SE splicing events had occurred during AS of the ABCB1 pre-mRNA that expresses ABCB1. This could explain the markedly reduced ABCB1 level in KB-C2-k.o.cdk6 cells and enhanced expression of ABCB1 or its isoform in KB-C2-k.o.cdk4 cells, which resulted in different efficacy on ABCB1-mediated MDR. Interestingly, a knockout experiment indicated that cdk6 was downregulated along with the knockout of cdk4 in KB-C2 cells. To our knowledge, it is the first time that the responsive AS occurrence in the pre-mRNA of ABCB1 was found to be induced by the deletion of cdk6, cdk4 or PI3K 110α/β subunits. This will surely have significance in reversing ABCB1-mediated MDR by modulating cdk6 or cdk4 gene expression.
CDK6 has been suggested to be a redundant homolog of CDK4 in the past [62]. For the first time, our study demonstrated that CDK6 and CDK4 may be two homologous proteins that differentially regulate ABCB1 expression in MDR KB-C2 cells. Indeed, our results indicated that the knockout of cdk6 reversed ABCB1-mediated MDR in KB-C2 cells (Fig. 2, Fig. 6). In this study, differential gene expression revealed that, along with the deletion of CDK6, a series of signaling pathways altered, for example, JUN/MAPK10 which is crucial in regulating tumorigenesis [42, 63], NOTCH3 which is involved in carcinoma and immune tolerance [64], were downregulated in KB-C2-k.o.cdk6 cell population, whereas CYP2D6 which was responsible for the metabolism of many drugs [65], was upregulated. Combining our findings that cdk6-deficient cancer cells lost ABCB1-mediated MDR via co-downregulating PI3K 110α/β, we deem that cdk6 could be a promising target for reversing ABCB1-mediated MDR and inhibiting cancer cell proliferation simultaneously. It may be hopeful to develop strategies based on targeting CDK6 for combined chemotherapy of MDR cancers with improved accuracy. In this aspect, modernized functional biomaterials can be integrated for further enhancing the targeting inhibition on CDK6 in cancer cells.
In summary, we demonstrated that regulation of ABCB1 can be realized through targeting CDK6-PI3K axis. Knockout of the cdk6 gene unveiled a novel targeting pathway for overcoming ABCB1-mediated MDR in cancers. PI3K 110α/β signaling was downregulated in the cdk6-deficient KB-C2 cell populations. A high frequency of responsive AS, mainly SE events, were induced by CDK6, CDK4, and PI3K 110α/β level changes, and these AS events finally led to the alteration of the ABCB1 expression and ABCB1-mediated MDR in cdk6 or cdk4 deficient KB-C2 cancer cells. Cross-regulation between CDK6 and PI3K110α/110β was discovered, and knockout of either one of these two genes could induce downregulation of the other and remarkable inhibition of ABCB1 expression, implying that they may synergize with each other to regulate ABCB1 expression. This will greatly strengthen the efficiency during reversing ABCB1 when using either of them as target.
With the knockout of cdk6, quite a few of signaling factors leading to inhibition of cancer cell proliferation were found to be promoted. For example, FOS-promoting cell autophagy [38] was upregulated. It is possible that these genes are quite variable in expression to adapt to CDK6 deficiency.
In the Western blot analysis, the positive signal band showed certain peptide with a reduced molecular weight in the cdk4 or cdk6 deficient cell populations. This should be induced by DNA repairment during CRISPR/Cas9 gene editing, that is, truncated proteins with similarity to wild-type CDK4 or CDK6 was produced after chromosomal repairment. To confirm this, we performed RT-PCR and certified deficiency of the objective region in cdk4 or cdk6 genes on chromosomes, and the results demonstrated knockout of wild-type cdk4 or cdk6 genes.
In vivo experiment demonstrated the function of cdk6 in maintaining ABCB1-mediated drug resistance in cancers. Since cdk6 deficiency or PIK3CA/PIK3CB deletion downregulated each other, and PIK3CA/PIK3CB knockout also downregulated ABCB1, we deem that CDK6-PI3K signaling axis is an efficient target for attenuating ABCB1-mediated MDR. To some degree, cdk6 deficiency inhibited KB-C2 tumor growth, which showed similar efficacy with DOX treated KB-C2 tumors. However, the efficacy was far less than the DOX-treated KB-C2-k.o.cdk6 tumor group. ABCB1-mediated drug resistance appeared to be a dominant barrier for inhibition on KB-C2 tumor.
Most known cancer cells have increased PIK3CA expression or more active PIK3CA which may lead to enhanced cell survivability, proliferative or anti-apoptosis ability, and overexpression of ABCB1 as we reported [2]. therefore, realizing downregulation of PIK3CA and ABCB1 by knocking out CDK6 in MDR cancer cells overexpressing ABCB1 might benefit combined chemotherapy against cancer cells with ABCB1-mediated MDR.
Through transcriptome analysis in this study, we found that protein CKAP4 is expressed at especially high level in both KB-C2 and KB-C2-k.o.cdk6 cells. It was reported that CKAP4 was minimally expressed in non-tumor cells [66], therefore, it was adopted as a biomarker for investigation of KB-C2 and KB-C2-k.o.cdk6 metastasis in the tissues of nude mice. We performed transcriptome sequencing and quantification to test the protein expression at the transcription level. The results reported that CDK6 expression in MDR KB-C2 cells (over expressing ABCB1, Fig. S4) was elevated to approximately 7 folds (Fig. S5) compared to drug sensitive parental KB-3-1 cells which did not express detectable amount of ABCB1 (Fig. S4). This provided a clue for clarifying the regulatory effect of CDK6 on ABCB1 by specific gene knockout experiment. By knocking down cdk6 in KB-C2 cell population, metastasis and survival of KB-C2-k.o.cdk6 tumor cells were remarkably inhibited in the DOX-treated nude mice, mainly because the drug resistance of KB-C2-k.o.cdk6 was inhibited compared with KB-C2.
Although a combination of prospective and retrospective cohort study involving 131 cases of advanced stage invasive breast cancer (which have received neoadjuvant chemotherapy) indicated that the expression of ABCB1 had no significant statistical correlation to metastases (p = 0.659) [67], whether ABCB1 has any effect on tumor cell migration, which has not been clearly studied, is an undergoing project we are studying.
Based on these findings, combined targeted chemotherapy with improved efficacy can be realized through downregulating CDK6-PI3K signaling axis in cancers overexpressing ABCB1, which is a common ABC member overexpressed in most malignant tumors and functions to generate drug resistance by efflux of its substance drugs.
Conclusion
In conclusion, we deem that CDK6-PI3K axis can be ideal target for reversing ABCB1 mediated MDR in cancers without inducing cancer cell proliferation, and this finding is of great significance for the combined anticancer chemotherapy which can reverse multidrug resistance and inhibit the growth of tumor cells at the same time. This study revealed the cross downregulation between CDK6 and PI3K in cdk6 deficient cancer cell populations, demonstrated that CDK6-PI3K axis could be new target for inhibiting ABCB1-mediated MDR. Meanwhile, knockout of cdk6 could inhibit cancer cell proliferation and malignancy. These new findings will surely benefit exploration of new drugs targeting cdk6 or PIK3CA/PIK3CB genes or gene products through which the therapeutic effect could be optimized.
Supplementary Information
Additional file 1: Fig. S1. Network and the interactions of the Top10 differentially expressed genes involved in KEGG Endocrine resistance. A Network of enriched terms colored by cluster ID, where nodes that share the same cluster ID are typically close to each other. B Protein-protein interaction network and components identified in the gene lists. String platform was used for the analysis.
Additional file 2: Fig. S2. Protein-protein interaction of the proteins which are most correlated with endocrine resistance.
Additional file 3: Fig. S3. Regulation of CDK6 on ABCB1-mediated MDR in H460/MX80. A RT-PCR certification of cdk6 or cdk4 gene knockout in H460/MX80 cells. The cells were transfected with the CRISPR/Cas9 all-in-one plasmid and stabilized for 2 weeks and then analyzed for the cdk4 or cdk6 mRNA amounts. B MTT analysis of cell viability of MDR H460/MX80 cells (with very low level of ABCB1 expression) and the cdk6-deficient H460/MX80 cells, i.e., H460/MX80-k.o.cdk6 cells. Paclitaxel and colchicine, which are ABCB1 substates, were used for co-culture with H460/MX80 and H460/MX80-k.o.cdk6 cells for 72 h.
Additional file 4: Fig. S4. Upregulation of ABCB1 in MDR KB-C2 cells compared to KB-3-1 cells. Transcriptome sequencing and quantification were performed in the MDR KB-C2 cells and drug sensitive parental KB-3-1 cells in the same condition, and the data showing the level of all the ABCB1 transcripts is summarized in the graph. As indicated by the transcripts per million mapped reads (TPM) value, the number of full-length-ABCB1 transcripts (mRNA) in KB-C2 cells were substantially increased as compared with that in KB-3-1 cells. The transcripts ENST00000265724, ENST00000622132 and MSTRG.27385.1 corresponding to the full-length of ABCB1 protein were indicated by *.
Additional file 5: Fig. S5. Upregulation of CDK6 in MDR KB-C2 cells compared to KB-3-1 cells. Transcriptome sequencing and quantification were performed in the MDR KB-C2 cells and drug sensitive parental KB-3-1 cells in the same condition, and the data showing the level of all the cdk6 transcripts is summarized in this graph. As indicated by the transcripts per million mapped reads (TPM) value, the number of full-length-cdk6 transcripts (mRNA) in KB-C2 cells were 7-folds of that in KB-3-1 cells. The transcripts ENST00000265734 and ENST00000424848 corresponding to the full-length of CDK6 protein were indicated by *.
Additional file 6: Table S1. Differential expression of CDK6, ABCB1, CDK4, PIK3CA, PIK3CB and GAPDH genes in KB-C2 and KB-C2-k.o.cdk6. Identical amounts of total RNA from KB-C2 and KB-C2-k.o.cdk6 cells were analyzed for transcriptome sequences and differential gene expression. The transcripts encoding full or effective length of proteins are listed.
Additional file 7: Table S2. Downregulation of the expression level of CDK6 in KB-C2-k.o.110α, KB-C2-k.o.110β, MX80-k.o.110α, or MX80-k.o.110β populations.
Additional file 8: Table S3. The percentages of non-apoptotic cells in the different phases of the cell cycle. The data were calculated based on the results in the histograms indicating a representative cell cycle (Fig. 6).
Acknowledgements
We would like to thank Charles R. Ashby (College of Pharmacy and Health Sciences, St. John’s University) for editorial assistance. We thank Dr. Shin-Ich Akiyama (Kagoshima University, Japan) for providing us the KB-3-1 and KB-C2 cell lines, Drs. Susan E. Bates and Robert W. Robey (NIH, Bethesda, MD) for providing us NCl-H460 cell line.
Abbreviations
- ABC transporter
ATP-binding cassette (ABC) transporter
- ACTN2
Alpha-actinin 2
- AF488
Alexa Fluor 488
- AGE
Advanced glycation end product
- AGT
Angiotensinogen
- AKT/PKB
Protein kinase B
- ATP2A2
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2
- ATP5ME
Mitochondrial ATP synthase subunit e
- ATP5PF
Mitochondrial ATP synthase-coupling factor 6
- ATP6V1B2
Brain V-type proton ATPase subunit B2
- AS
Alternative splicing
- BCRP/ABCG2
Breast cancer resistance protein
- CCNA2
Cyclin A2 gene
- MET
Mesenchymal to epithelial transition factor, a hepatocyte growth factor receptor tyrosine kinase
- CDKN2B
Cyclin dependent kinase inhibitor 2B
- CDKN2C
Cyclin dependent kinase inhibitor 2C
- CDK4
Cyclin dependent kinase 4
- cdk4
Gene of CDK4
- CDK6
Cyclin dependent kinase 6
- cdk6
Gene of CDK6
- CDKs
Cyclin dependent kinases
- CD36
Cluster of differentiation 36
- CKAP4
Cytoskeleton-associated protein 4
- CRISPR
Clustered regularly interspaced short palindromic repeats
- CXCL1
Chemokine (C-X-C motif) ligand 1
- CYP2D6
Cytochrome P450 family 2 subfamily D member 6
- DAPI
4′,6-diamidino-2-phenylindole
- DEG
Differential expression genes
- DMSO
Dimethyl sulfoxide
- EC50
Concentration for 50% of maximal effect
- EGFR
Epidermal growth factor receptor
- EMT
Epithelial-mesenchymal transition
- ERK
Extracellular regulated protein kinase
- E2F
E2 transcription factor
- FAT1
FAT atypical cadherin 1
- FBS
Fatal bovine serum
- FDA
Food and Drug Administration (FDA)
- FGF11
Fibroblast growth factor 11
- FGF18
Fibroblast growth factor 18
- FGF21
Fibroblast growth factor 21
- FLT3-ITD
Fms-like tyrosine kinase 3 - in-frame internal tandem duplications
- FN1
Fibronectin 1
- FOS
Fos proto-oncogene, AP-1 transcription factor subunit
- FOXO3
Forkhead box O3
- FRKM
Fragments per kilobase of exon per million mapped reads
- GADD45A
Growth arrest and DNA damage inducible alpha
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- GDF6
Growth differentiation factor 6
- GLI2
GLI family zinc finger 2
- GPER1
G-protein coupled estrogen receptor 1
- HCK
SRC family kinase HCK
- HRP
Horseradish peroxidase
- IC50
50% inhibiting concentration
- IF
Immune-fluorescence
- INHBA
Inhibin subunit beta A
- ITGA11
Integrin subunit alpha 11
- JNK
c-Jun N-terminal kinase
- JUN
Jun proto-oncogene, AP-1 transcription factor subunit
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MAP K10
Mitogen-activated protein kinase 10
- MATS
Multivariate analysis of transcript splicing
- MDR
Multidrug resistance
- MEK
Mitogen-activated extracellular signal-regulated kinase
- miRNA
micro RNA
- MYH7
Myosin heavy chain 7
- NBDs
Nucleotide-binding domains
- Neu2
Cytosolic sialidase
- NF-κB
Facilitates nuclear factor kappaB
- NGFR
Nerve growth factor receptor
- NOTCH3
Neurogenic locus notch homolog protein 3
- NOTCH2NLB
Notch 2 N-terminal like B
- ORF
Open reading frame
- PDPK1
3-phosphoinositide dependent protein kinase 1
- P-gp
P-glycoprotein
- PI3K
Phosphoinositide 3-kinase
- P110α
PI3K subunits PIK3CA
- P110β
PI3K subunits PIK3CB
- Raf
Rapidly accelerated Fibrosarcoma
- PIK3R2
Phosphatidylinositol 3-kinase regulatory subunit beta
- PIK3R3
Phosphatidylinositol 3-kinase regulatory subunit gamma
- PKA
Protein kinase A
- PPI
Protein-protein interaction
- PRKACB
cAMP-dependent protein kinase catalytic subunit beta
- PTGS2
Prostaglandin-endoperoxide synthase 2
- RAGE
AGE receptor
- Ras
Rat sarcoma
- RASSF5
Ras association domain family member 5
- Rb
Retinoblastoma protein
- ROS
Reactive oxygen species
- SE
Skipped exon
- SgRNA
Single guide RNA
- SERCA
Sarco/endoplasmic reticulum Ca2+-ATPases
- SERPINE1
Serpin family E member 1
- SOX2
SPY homology box 2
- TGFA
Transforming growth factor alpha
- TGFBR1
Transforming growth factor beta receptor 1
- TMDs
Transmembrane domains
- TPM
Transcripts per million mapped reads
Authors’ contributions
LZ and ZSC conceived this study and designed the experiments. LZ, ZSC, JZ, SF provided direction and guidance on the whole project. LZ, CH, JZ, SF, ZC, and ZSC sponsored the project. LZ, YL performed Western blot, MTT, and immunofluorescence analysis, LZ performed gene knockout, the bioinformatic analysis, the cell cycle analysis and in vivo studies, LZ wrote the paper, LZ, ZSC, YC, CH, JZ, SF, and ZC revised the paper. All authors read and approved the final manuscript.
Funding
This work was funded by the National Natural Science Foundation of China (21877113, 81572944), the Natural Science Foundation of Fujian Province (2020I0036), Guangdong Basic and Applied Basic Research Regional Combination The Youth Foundation (2019A1515110155), Guangdong Natural Science Foundation Surface Project (2021A1515010807, 2020A1515010605), Shenzhen Natural Science Foundation Basic Research Surface Project (JCYJ20210324123012035), the National Key Research and Development Program (2018YFA0902801) and the Startup Fund for the 100 Top Talents Program, SYSU (392001). This study was partially supported by the Department of Pharmaceutical Sciences, College of Pharmacy and Health Sciences, St. John’s University (New York, USA).
Availability of data and materials
Sequence information of cdk4 (NM_000075.3) and cdk6 (NM_001259.7) are available in GenBank. All other data are available in the main text or the supplementary materials. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors declare no competing interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Lei Zhang, Email: zhangl@fjirsm.ac.cn.
Jian-Ye Zhang, Email: jianyez@163.com.
Shuo Fang, Email: fangsh9@mail.sysu.edu.cn.
References
- 1.Kathawala RJ, Gupta P, Ashby CR, Chen ZS. The modulation of ABC transporter-mediated multidrug resistance in cancer: a review of the past decade. Drug Resist Update. 2015;18:1–17. doi: 10.1016/j.drup.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 2.Zhang L, Li Y, Wang Q, Chen Z, Li X, Wu Z, et al. The PI3K subunits, P110alpha and P110beta are potential targets for overcoming P-gp and BCRP-mediated MDR in cancer. Mol Cancer. 2020;19:10. doi: 10.1186/s12943-019-1112-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loyer P, Trembley JH. Roles of CDK/cyclin complexes in transcription and pre-mRNA splicing: cyclins L and CDK11 at the cross-roads of cell cycle and regulation of gene expression. Semin Cell Dev Biol. 2020;107:36–45. doi: 10.1016/j.semcdb.2020.04.016. [DOI] [PubMed] [Google Scholar]
- 4.Liang S, Hu L, Wu Z, Chen Z, Liu S, Xu X, et al. CDK12: a potent target and biomarker for human cancer therapy. Cells. 2020;9:1483. doi: 10.3390/cells9061483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robert T, Johnson JL, Guichaoua R, Yaron TM, Bach S, Cantley LC, et al. Development of a CDK10/CycM in vitro kinase screening assay and identification of first small-molecule inhibitors. Front Chem. 2020;8:147. doi: 10.3389/fchem.2020.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Czudor Z, Balogh M, Banhegyi P, Boros S, Breza N, Dobos J, et al. Novel compounds with potent CDK9 inhibitory activity for the treatment of myeloma. Bioorg Med Chem Lett. 2018;28:769–773. doi: 10.1016/j.bmcl.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 7.Menzl I, Zhang T, Berger-Becvar A, Grausenburger R, Heller G, Prchal-Murphy M, et al. A kinase-independent role for CDK8 in BCR-ABL1(+) leukemia. Nat Commun. 2019;10:4741. doi: 10.1038/s41467-019-12656-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rainey MD, Quachthithu H, Gaboriau D, Santocanale C. DNA replication dynamics and cellular responses to ATP competitive CDC7 kinase inhibitors. ACS Chem Biol. 2017;12:1893–1902. doi: 10.1021/acschembio.7b00117. [DOI] [PubMed] [Google Scholar]
- 9.Duan C, Liu Y, Lu L, Cai R, Xue H, Mao X, et al. CDK14 contributes to reactive gliosis via interaction with cyclin Y in rat model of spinal cord injury. J Mol Neurosci. 2015;57:571–579. doi: 10.1007/s12031-015-0639-x. [DOI] [PubMed] [Google Scholar]
- 10.Li Q, Liu X, Zhang M, Ye G, Qiao Q, Ling Y, et al. Characterization of a novel human CDK5 splicing variant that inhibits Wnt/beta-catenin signaling. Mol Biol Rep. 2010;37:2415–2421. doi: 10.1007/s11033-009-9752-7. [DOI] [PubMed] [Google Scholar]
- 11.Hirai H, Shimomura T, Kobayashi M, Eguchi T, Taniguchi E, Fukasawa K, et al. Biological characterization of 2-aminothiazole-derived Cdk4/6 selective inhibitor in vitro and in vivo. Cell Cycle. 2010;9:1590–1600. doi: 10.4161/cc.9.8.11306. [DOI] [PubMed] [Google Scholar]
- 12.Braun K, Holzl G, Pusch O, Hengstschlager M. Deregulated expression of CDK2- or CDK3-associated kinase activities enhances c-Myc-induced apoptosis. DNA Cell Biol. 1998;17:789–798. doi: 10.1089/dna.1998.17.789. [DOI] [PubMed] [Google Scholar]
- 13.Vanstraelen M, Torres Acosta JA, De Veylder L, Inze D, Geelen D. A plant-specific subclass of C-terminal kinesins contains a conserved a-type cyclin-dependent kinase site implicated in folding and dimerization. Plant Physiol. 2004;135:1417–1429. doi: 10.1104/pp.104.044818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schymkowitz JW, Rousseau F, Itzhaki LS. Sequence conservation provides the best prediction of the role of proline residues in p13suc1. J Mol Biol. 2000;301:199–204. doi: 10.1006/jmbi.2000.3958. [DOI] [PubMed] [Google Scholar]
- 15.Gray N, Detivaud L, Doerig C, Meijer L. ATP-site directed inhibitors of cyclin-dependent kinases. Curr Med Chem. 1999;6:859–875. doi: 10.2174/092986730609220401152358. [DOI] [PubMed] [Google Scholar]
- 16.Swaffer MP, Jones AW, Flynn HR, Snijders AP, Nurse P. CDK substrate phosphorylation and ordering the cell cycle. Cell. 2016;167:1750–1761 e1716. doi: 10.1016/j.cell.2016.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu H, Yu S, Liu Q, Yuan X, Mani S, Pestell RG, et al. Recent advances of highly selective CDK4/6 inhibitors in breast cancer. J Hematol Oncol. 2017;10:97. doi: 10.1186/s13045-017-0467-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bates S, Parry D, Bonetta L, Vousden K, Dickson C, Peters G. Absence of cyclin D/cdk complexes in cells lacking functional retinoblastoma protein. Oncogene. 1994;9:1633–1640. [PubMed] [Google Scholar]
- 19.Guan KL, Jenkins CW, Li Y, Nichols MA, Wu X, O'Keefe CL, et al. Growth suppression by p18, a p16INK4/MTS1- and p14INK4B/MTS2-related CDK6 inhibitor, correlates with wild-type pRb function. Genes Dev. 1994;8:2939–2952. doi: 10.1101/gad.8.24.2939. [DOI] [PubMed] [Google Scholar]
- 20.VanArsdale T, Boshoff C, Arndt KT, Abraham RT. Molecular pathways: targeting the cyclin D-CDK4/6 axis for cancer treatment. Clin Cancer Res. 2015;21:2905–2910. doi: 10.1158/1078-0432.CCR-14-0816. [DOI] [PubMed] [Google Scholar]
- 21.Nath S, Mandal C, Chatterjee U, Mandal C. Association of cytosolic sialidase Neu2 with plasma membrane enhances Fas-mediated apoptosis by impairing PI3K-Akt/mTOR-mediated pathway in pancreatic cancer cells. Cell Death Dis. 2018;9:210. doi: 10.1038/s41419-017-0191-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Z, Razavi P, Li Q, Toy W, Liu B, Ping C, et al. Loss of the FAT1 tumor suppressor promotes resistance to CDK4/6 inhibitors via the hippo pathway. Cancer Cell. 2018;34:893–905 e898. doi: 10.1016/j.ccell.2018.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, Ge X, Zhang W, Ding P, Du Y, Wang Q, et al. PI3K/AKT inhibition reverses R-CHOP resistance by destabilizing SOX2 in diffuse large B cell lymphoma. Theranostics. 2020;10:3151–3163. doi: 10.7150/thno.41362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, Fan C, Pu L, Wei C, Jin H, Teng Y, et al. Phloretin induces cell cycle arrest and apoptosis of human glioblastoma cells through the generation of reactive oxygen species. J Neuro-Oncol. 2016;128:217–223. doi: 10.1007/s11060-016-2107-z. [DOI] [PubMed] [Google Scholar]
- 25.Li J, Ying Y, Xie H, Jin K, Yan H, Wang S, et al. Dual regulatory role of CCNA2 in modulating CDK6 and MET-mediated cell-cycle pathway and EMT progression is blocked by miR-381-3p in bladder cancer. FASEB J. 2019;33:1374–1388. doi: 10.1096/fj.201800667R. [DOI] [PubMed] [Google Scholar]
- 26.Uras IZ, Maurer B, Nivarthi H, Jodl P, Kollmann K, Prchal-Murphy M, et al. CDK6 coordinates JAK2 (V617F) mutant MPN via NF-kappaB and apoptotic networks. Blood. 2019;133:1677–1690. doi: 10.1182/blood-2018-08-872648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu QL, Xu ZP, Lan YF, Li B. miR-636 represses cell survival by targeting CDK6/Bcl-2 in cervical cancer. Kaohsiung J Med Sci. 2020;36:328–335. doi: 10.1002/kjm2.12181. [DOI] [PubMed] [Google Scholar]
- 28.Liu Z, Long X, Chao C, Yan C, Wu Q, Hua S, et al. Knocking down CDK4 mediates the elevation of let-7c suppressing cell growth in nasopharyngeal carcinoma. BMC Cancer. 2014;14:274. doi: 10.1186/1471-2407-14-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sorf A, Sucha S, Morell A, Novotna E, Staud F, Zavrelova A, et al. Targeting pharmacokinetic drug resistance in acute myeloid leukemia cells with CDK4/6 inhibitors. Cancers (Basel) 2020;12:1596. doi: 10.3390/cancers12061596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sumi NJ, Kuenzi BM, Knezevic CE, Remsing Rix LL, Rix U. Chemoproteomics reveals novel protein and lipid kinase targets of clinical CDK4/6 inhibitors in lung Cancer. ACS Chem Biol. 2015;10:2680–2686. doi: 10.1021/acschembio.5b00368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinez-Chavez A, van Hoppe S, Rosing H, Lebre MC, Tibben M, Beijnen JH, et al. P-glycoprotein limits ribociclib brain exposure and CYP3A4 restricts its oral bioavailability. Mol Pharm. 2019;16:3842–3852. doi: 10.1021/acs.molpharmaceut.9b00475. [DOI] [PubMed] [Google Scholar]
- 32.Akiyama S, Fojo A, Hanover JA, Pastan I, Gottesman MM. Isolation and genetic characterization of human KB cell lines resistant to multiple drugs. Somat Cell Mol Genet. 1985;11:117–126. doi: 10.1007/BF01534700. [DOI] [PubMed] [Google Scholar]
- 33.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson MD, McCarthy DJ. Smyth GK: edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen S, Park JW, Huang J, Dittmar KA, Lu ZX, Zhou Q, Carstens RP, Xing Y. MATS: a Bayesian framework for flexible detection of differential alternative splicing from RNA-Seq data. Nucleic Acids Res. 2012;40:e61. doi: 10.1093/nar/gkr1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oh JM, Kim E, Chun S. Ginsenoside compound K induces Ros-mediated apoptosis and Autophagic inhibition in human neuroblastoma cells in vitro and in vivo. Int J Mol Sci. 2019;20:4279. [DOI] [PMC free article] [PubMed]
- 38.Shao J, Jiang F, Hu M, Mei E, Pan Z, Chen C, et al. The role of FOS-mediated autophagy activation in the indocyanine green-based photodynamic therapy for treating melanoma. J Photochem Photobiol B. 2021;214:112101. doi: 10.1016/j.jphotobiol.2020.112101. [DOI] [PubMed] [Google Scholar]
- 39.Tsuchida N, Ryder T, Ohtsubo E. Nucleotide sequence of the oncogene encoding the p21 transforming protein of Kirsten murine sarcoma virus. Science. 1982;217:937–939. doi: 10.1126/science.6287573. [DOI] [PubMed] [Google Scholar]
- 40.Thein KZ, Biter AB, Hong DS. Therapeutics targeting mutant KRAS. Annu Rev Med. 2021;72:349–364. doi: 10.1146/annurev-med-080819-033145. [DOI] [PubMed] [Google Scholar]
- 41.Vogt PK. Fortuitous convergences: the beginnings of JUN. Nat Rev Cancer. 2002;2:465–469. doi: 10.1038/nrc818. [DOI] [PubMed] [Google Scholar]
- 42.Sun R, Xiang T, Tang J, Peng W, Luo J, Li L, et al. 19q13 KRAB zinc-finger protein ZNF471 activates MAPK10/JNK3 signaling but is frequently silenced by promoter CpG methylation in esophageal cancer. Theranostics. 2020;10:2243–2259. doi: 10.7150/thno.35861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qiao B, Wang Q, Zhao Y, Wu J. miR-205-3p functions as a tumor suppressor in ovarian carcinoma. Reprod Sci. 2020;27:380–388. doi: 10.1007/s43032-019-00047-y. [DOI] [PubMed] [Google Scholar]
- 44.Pankov R, Yamada KM. Fibronectin at a glance. J Cell Sci. 2002;115:3861–3863. doi: 10.1242/jcs.00059. [DOI] [PubMed] [Google Scholar]
- 45.Cai X, Liu C, Zhang TN, Zhu YW, Dong X, Xue P. Down-regulation of FN1 inhibits colorectal carcinogenesis by suppressing proliferation, migration, and invasion. J Cell Biochem. 2018;119:4717–4728. doi: 10.1002/jcb.26651. [DOI] [PubMed] [Google Scholar]
- 46.Xi X, Liu N, Wang Q, Chu Y, Yin Z, Ding Y, et al. ACT001, a novel PAI-1 inhibitor, exerts synergistic effects in combination with cisplatin by inhibiting PI3K/AKT pathway in glioma. Cell Death Dis. 2019;10:757. doi: 10.1038/s41419-019-1986-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prasad V, Okunade GW, Miller ML, Shull GE. Phenotypes of SERCA and PMCA knockout mice. Biochem Biophys Res Commun. 2004;322:1192–1203. doi: 10.1016/j.bbrc.2004.07.156. [DOI] [PubMed] [Google Scholar]
- 48.Dang D, Rao R. Calcium-ATPases: gene disorders and dysregulation in cancer. Biochim Biophys Acta. 1863;2016:1344–1350. doi: 10.1016/j.bbamcr.2015.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gamm DM, Baude EJ, Uhler MD. The major catalytic subunit isoforms of cAMP-dependent protein kinase have distinct biochemical properties in vitro and in vivo. J Biol Chem. 1996;271:15736–15742. doi: 10.1074/jbc.271.26.15736. [DOI] [PubMed] [Google Scholar]
- 50.Chen Y, Gao Y, Tian Y, Tian DL. PRKACB is downregulated in non-small cell lung cancer and exogenous PRKACB inhibits proliferation and invasion of LTEP-A2 cells. Oncol Lett. 2013;5:1803–1808. doi: 10.3892/ol.2013.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou J, Chen GB, Tang YC, Sinha RA, Wu Y, Yap CS, et al. Genetic and bioinformatic analyses of the expression and function of PI3K regulatory subunit PIK3R3 in an Asian patient gastric cancer library. BMC Med Genet. 2012;5:34. doi: 10.1186/1755-8794-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vallejo-Diaz J, Chagoyen M, Olazabal-Moran M, Gonzalez-Garcia A, Carrera AC. The opposing roles of PIK3R1/p85alpha and PIK3R2/p85beta in Cancer. Trends Cancer. 2019;5:233–244. doi: 10.1016/j.trecan.2019.02.009. [DOI] [PubMed] [Google Scholar]
- 53.Vries MH, Wagenaar A, Verbruggen SE, Molin DG, Dijkgraaf I, Hackeng TH, et al. CXCL1 promotes arteriogenesis through enhanced monocyte recruitment into the peri-collateral space. Angiogenesis. 2015;18:163–171. doi: 10.1007/s10456-014-9454-1. [DOI] [PubMed] [Google Scholar]
- 54.Hsu YL, Chen YJ, Chang WA, Jian SF, Fan HL, Wang JY, et al. Interaction between tumor-associated dendritic cells and Colon Cancer cells contributes to tumor progression via CXCL1. Int J Mol Sci. 2018;19:2427. [DOI] [PMC free article] [PubMed]
- 55.Gigant B, Cormier A, Dorleans A, Ravelli RB, Knossow M. Microtubule-destabilizing agents: structural and mechanistic insights from the interaction of colchicine and vinblastine with tubulin. Top Curr Chem. 2009;286:259–278. doi: 10.1007/128_2008_11. [DOI] [PubMed] [Google Scholar]
- 56.Dallavalle S, Dobricic V, Lazzarato L, Gazzano E, Machuqueiro M, Pajeva I, et al. Improvement of conventional anti-cancer drugs as new tools against multidrug resistant tumors. Drug Resist Updat. 2020;50:100682. doi: 10.1016/j.drup.2020.100682. [DOI] [PubMed] [Google Scholar]
- 57.Mollazadeh S, Sahebkar A, Hadizadeh F, Behravan J, Arabzadeh S. Structural and functional aspects of P-glycoprotein and its inhibitors. Life Sci. 2018;214:118–123. doi: 10.1016/j.lfs.2018.10.048. [DOI] [PubMed] [Google Scholar]
- 58.Ji N, Yang YQ, Lei ZN, Cai CY, Wang JQ, Gupta P, et al. Ulixertinib (BVD-523) antagonizes ABCB1-and ABCG2-mediated chemotherapeutic drug resistance. Biochem Pharmacol. 2018;158:274–285. doi: 10.1016/j.bcp.2018.10.028. [DOI] [PubMed] [Google Scholar]
- 59.Zhang Y, Guo L, Huang J, Sun Y, He F, Zloh M, et al. Inhibitory effect of berberine on broiler P-glycoprotein expression and function: in situ and in vitro studies. Int J Mol Sci. 2019;20:1966. [DOI] [PMC free article] [PubMed]
- 60.Zhang QC, Song YG, Cheng XS, Xu ZW, Matthew OA, Wang J, et al. Apatinib reverses paclitaxel-resistant lung Cancer cells (A549) through blocking the function of ABCB1 transporter. Anticancer Res. 2019;39:5461–5471. doi: 10.21873/anticanres.13739. [DOI] [PubMed] [Google Scholar]
- 61.Gao X, Leone GW, Wang H. Cyclin D-CDK4/6 functions in cancer. Adv Cancer Res. 2020;148:147–169. doi: 10.1016/bs.acr.2020.02.002. [DOI] [PubMed] [Google Scholar]
- 62.Tigan AS, Bellutti F, Kollmann K, Tebb G, Sexl V. CDK6-a review of the past and a glimpse into the future: from cell-cycle control to transcriptional regulation. Oncogene. 2016;35:3083–3091. doi: 10.1038/onc.2015.407. [DOI] [PubMed] [Google Scholar]
- 63.Cui R, Jiang N, Zhang M, Du S, Ou H, Ge R, et al. AMOTL2 inhibits JUN Thr239 dephosphorylation by binding PPP2R2A to suppress the proliferation in non-small cell lung cancer cells. Biochim Biophys Acta Mol Cell Res. 1868;2021:118858. doi: 10.1016/j.bbamcr.2020.118858. [DOI] [PubMed] [Google Scholar]
- 64.Cui Y, Li Q, Li W, Wang Y, Lv F, Shi X, et al. NOTCH3 is a prognostic factor and is correlated with immune tolerance in gastric Cancer. Front Oncol. 2020;10:574937. doi: 10.3389/fonc.2020.574937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tolledo C, Stocco MR, Miksys S, Gonzalez FJ, Tyndale RF. Human CYP2D6 is functional in brain in vivo: evidence from humanized CYP2D6 transgenic mice. Mol Neurobiol. 2020;57:2509–2520. doi: 10.1007/s12035-020-01896-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shinno N, Kimura H, Sada R, Takiguchi S, Mori M, Fumoto K, Doki Y, Kikuchi A. Activation of the Dickkopf1-CKAP4 pathway is associated with poor prognosis of esophageal cancer and anti-CKAP4 antibody may be a new therapeutic drug. Oncogene. 2018;37:3471–3484. doi: 10.1038/s41388-018-0179-2. [DOI] [PubMed] [Google Scholar]
- 67.Prajoko YW, Aryandono T. The effect of P-glycoprotein (P-gp), nuclear factor-kappa B (Nf-kappab), and aldehyde Dehydrogenase-1 (ALDH-1) expression on metastases, recurrence and survival in advanced breast Cancer patients. Asian Pac J Cancer Prev. 2019;20:1511–1518. doi: 10.31557/APJCP.2019.20.5.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1. Network and the interactions of the Top10 differentially expressed genes involved in KEGG Endocrine resistance. A Network of enriched terms colored by cluster ID, where nodes that share the same cluster ID are typically close to each other. B Protein-protein interaction network and components identified in the gene lists. String platform was used for the analysis.
Additional file 2: Fig. S2. Protein-protein interaction of the proteins which are most correlated with endocrine resistance.
Additional file 3: Fig. S3. Regulation of CDK6 on ABCB1-mediated MDR in H460/MX80. A RT-PCR certification of cdk6 or cdk4 gene knockout in H460/MX80 cells. The cells were transfected with the CRISPR/Cas9 all-in-one plasmid and stabilized for 2 weeks and then analyzed for the cdk4 or cdk6 mRNA amounts. B MTT analysis of cell viability of MDR H460/MX80 cells (with very low level of ABCB1 expression) and the cdk6-deficient H460/MX80 cells, i.e., H460/MX80-k.o.cdk6 cells. Paclitaxel and colchicine, which are ABCB1 substates, were used for co-culture with H460/MX80 and H460/MX80-k.o.cdk6 cells for 72 h.
Additional file 4: Fig. S4. Upregulation of ABCB1 in MDR KB-C2 cells compared to KB-3-1 cells. Transcriptome sequencing and quantification were performed in the MDR KB-C2 cells and drug sensitive parental KB-3-1 cells in the same condition, and the data showing the level of all the ABCB1 transcripts is summarized in the graph. As indicated by the transcripts per million mapped reads (TPM) value, the number of full-length-ABCB1 transcripts (mRNA) in KB-C2 cells were substantially increased as compared with that in KB-3-1 cells. The transcripts ENST00000265724, ENST00000622132 and MSTRG.27385.1 corresponding to the full-length of ABCB1 protein were indicated by *.
Additional file 5: Fig. S5. Upregulation of CDK6 in MDR KB-C2 cells compared to KB-3-1 cells. Transcriptome sequencing and quantification were performed in the MDR KB-C2 cells and drug sensitive parental KB-3-1 cells in the same condition, and the data showing the level of all the cdk6 transcripts is summarized in this graph. As indicated by the transcripts per million mapped reads (TPM) value, the number of full-length-cdk6 transcripts (mRNA) in KB-C2 cells were 7-folds of that in KB-3-1 cells. The transcripts ENST00000265734 and ENST00000424848 corresponding to the full-length of CDK6 protein were indicated by *.
Additional file 6: Table S1. Differential expression of CDK6, ABCB1, CDK4, PIK3CA, PIK3CB and GAPDH genes in KB-C2 and KB-C2-k.o.cdk6. Identical amounts of total RNA from KB-C2 and KB-C2-k.o.cdk6 cells were analyzed for transcriptome sequences and differential gene expression. The transcripts encoding full or effective length of proteins are listed.
Additional file 7: Table S2. Downregulation of the expression level of CDK6 in KB-C2-k.o.110α, KB-C2-k.o.110β, MX80-k.o.110α, or MX80-k.o.110β populations.
Additional file 8: Table S3. The percentages of non-apoptotic cells in the different phases of the cell cycle. The data were calculated based on the results in the histograms indicating a representative cell cycle (Fig. 6).
Data Availability Statement
Sequence information of cdk4 (NM_000075.3) and cdk6 (NM_001259.7) are available in GenBank. All other data are available in the main text or the supplementary materials. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.