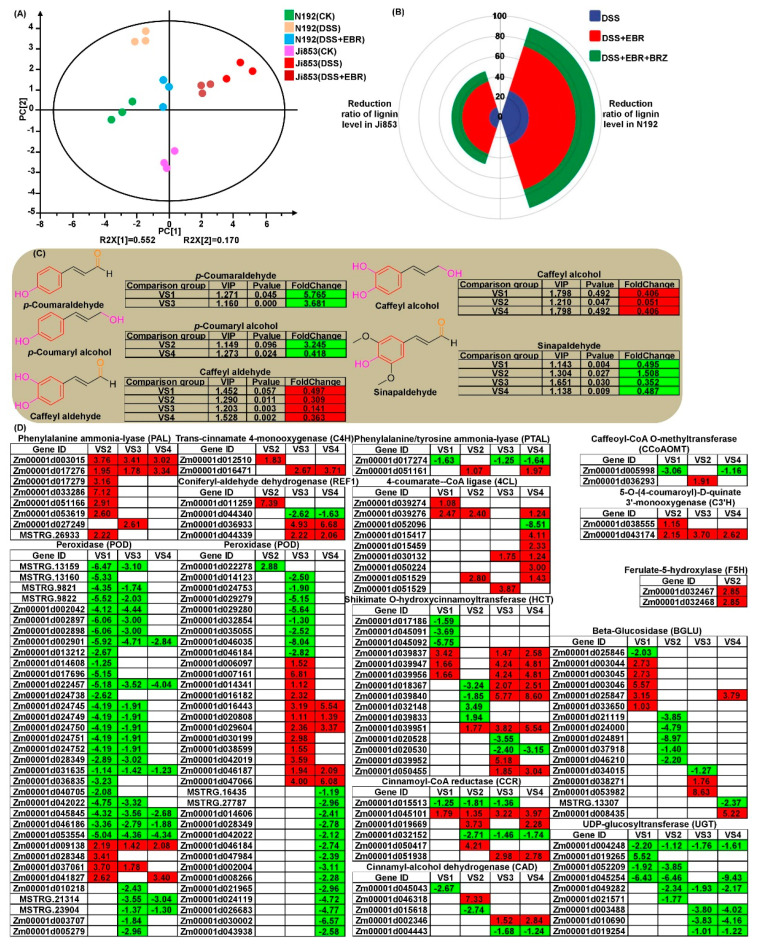
Figure 2.
Differentially expressed genes (DEGs) and differentially accumulated metabolites (DAMs) involved in lignin biosynthesis in N192 and Ji853 mesocotyls under CK, DSS, and DSS + EBR conditions. Principle component analysis (PCA) of the lignin metabolites derived from ultra–high–performance liquid chromatography-tandem mass spectrometry (UPLC–MS/MS) (A). Polar coordinate graph for reduction ratio of lignin level in N192 and Ji853 mesocotyls under DSS, DSS + EBR, and DSS + EBR + BRZ conditions that compared to CK control (B). DAMs were analyzed in all four comparisons [N192 (DSS_VS_CK), VS1], [N192 (DSS + EBR_VS_CK), VS2], [Ji853 (DSS_VS_CK), VS3], and [Ji853 (DSS + EBR_VS_CK), VS4]). The red and green box represented up– and down–accumulated DAMs, and the value in the box was the fold–change of DAMs (C). DEGs involved in lignin biosynthesis in four comparisons. The red and green box represented up– and down–regulated DEGs, and the value in the box was the log2 fold-change of DEGs (D).