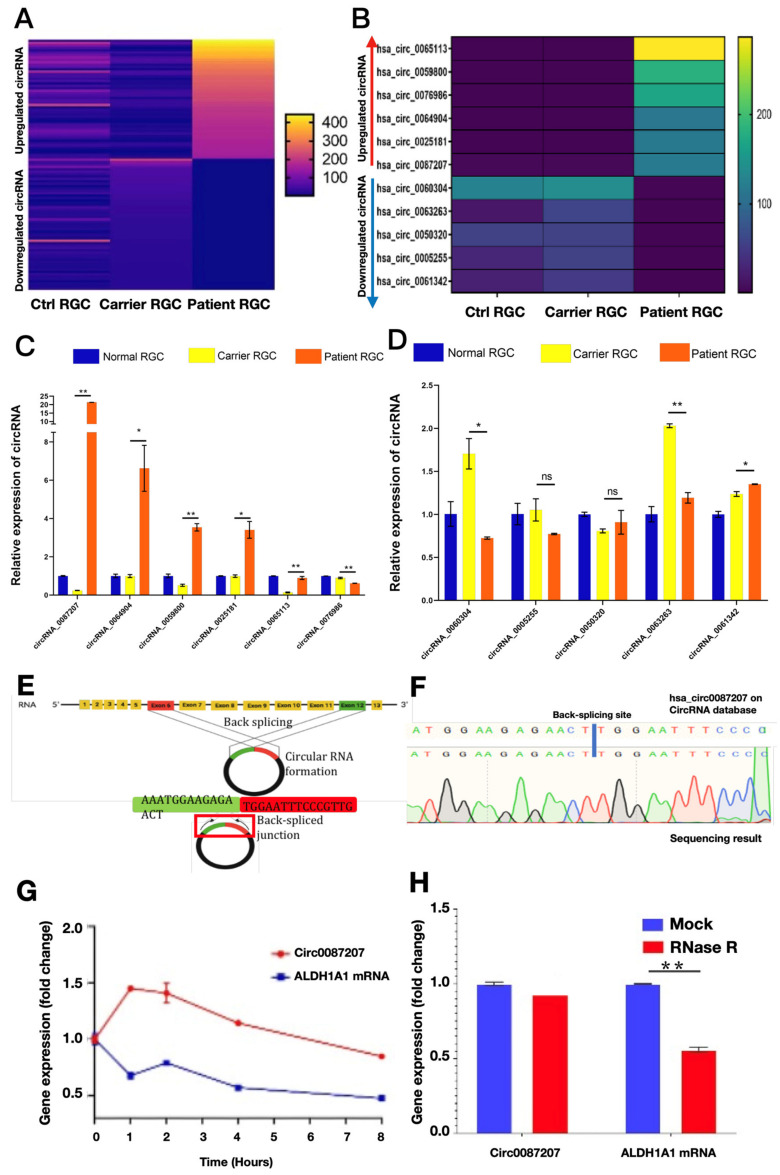
Figure 4.
Identification of upregulated circular RNAs in LHON patient iPSC-derived RGCs. (A) Heatmap analysis showed the circRNA profiling among the iPSC-derived RGCs generated from the control healthy subject, the carrier, and the LHON patient. (B) The top six upregulated circRNA and the top five downregulated circRNAs in LHON patient iPSC-derived RGCs were picked up and shown. The circRNA expression levels of the LHON patient iPSC-derived RGCs were compared with that of the carrier iPSC-derived RGCs. The next-generation sequencing results of (C) the most upregulated circRNAs and (D) the most downregulated circRNAs in LHON patient iPSC-derived RGCs were validated using quantitative real-time PCR. Among all the upregulated circRNAs, hsa_circ_0087207 exhibited the highest expression in the LHON patient iPSC-derived RGCs as compared with the carrier iPSC-derived RGCs. (E) The schematic diagram presented the formation of circ0087207 from the 6th and 12th exons of ALDH1A1 gene. (F) The sequence of the identified circRNA with the highest expression matched the sequence and back-splicing junction site of hsa_circ_0087207 published in the online circRNA library on Circular RNA Interactome. (G) Actinomycin D assay and (H) RNase R assay were used to examine the stability of hsa_circ_0087207. * p < 0.05; ** p < 0.01. ns p > 0.05.