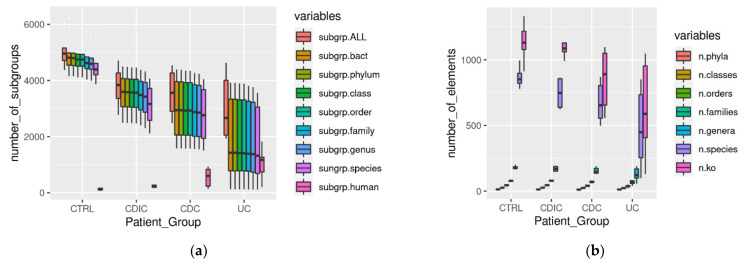
Figure 2.
(a) Protein subgroups (abbreviated ‘subgrp’) per sample in each clinical group: subgr.ALL: total number of protein subgroups; subgrp.bact: total number of bacterial subgroups; subgrp.phylum to subgrp.species: total number of bacterial subgroups with consensual taxonomic annotation of all their protein elements from the phylum-to-species level; subgrp.ko: number of bacterial subgroups with consensual functional annotation of all their protein elements; subgrp.human: total number of human subgroups. (b) n.phyla to n.species: number of phyla, classes,… species per sample (only taxonomic annotations concordant within a same protein subgroup were counted); n.ko: number of KO entries per ample (only functional annotations concordant within a same protein subgroup were counted). Abbreviations and headcounts of patient groups are those detailed in legend of Figure 1.