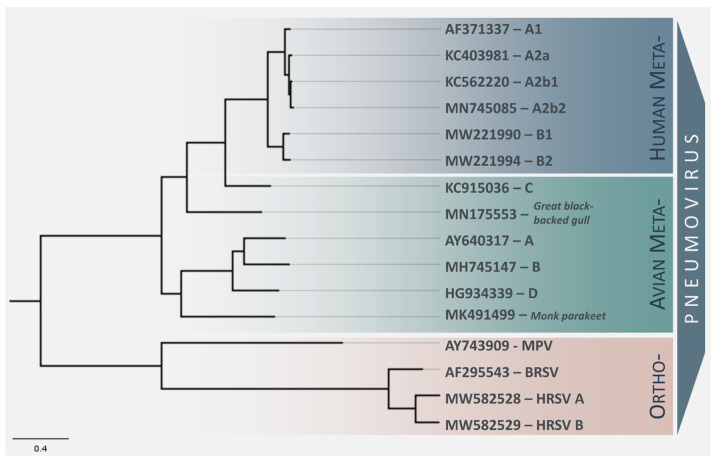
Figure 1.
Maximum likelihood phylogeny of metapneumovirus sequences. The phylogenetic tree was constructed using complete genome sequences from NCBI GenBank representing the different subgroups of AMPV and HMPV with orthopneumovirus sequences used as an outgroup. Maximum likelihood phylogeny was performed using a GTR + G model of nucleotide substitution. The tree was generated using bootstrap support of 1000 replications. The tip labels represent the NCBI GenBank accession numbers of respective virus strains, and the scale bar is proportional to the number of nucleotide substitutions per site.