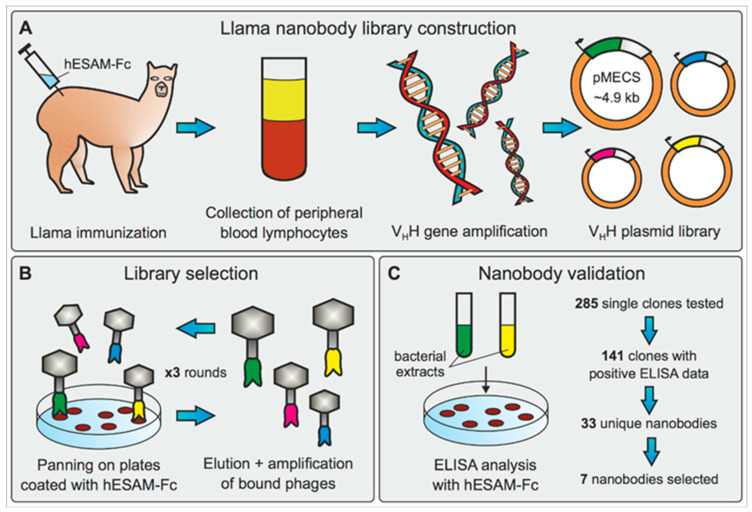
Figure 1.
Generation and identification of hESAM-specific Nbs. (A) Generation of hESAM-specific Nb phagemid library by llama immunization with a recombinant hESAM-Fc antigen, followed by collection of llama peripheral blood lymphocyte total RNA and subsequent VHH gene amplification. Resulting VHH gene products were integrated into pMECS phagemid vectors to obtain a VHH gene plasmid library. (B) For selection of hESAM-specific Nbs, the phagemid library was subjected to three consecutive rounds of biopanning on plates coated with the hESAM-Fc immunogen. After each round, bound phages were eluted and amplified in E. coli cells. (C) Binding specificity of enriched candidates was assessed by ELISA screening using bacterial extracts of 285 randomly selected clones. Out of 141 positive binders, sequence analysis identified 33 unique Nbs, and 7 Nb clones were selected for further analysis.