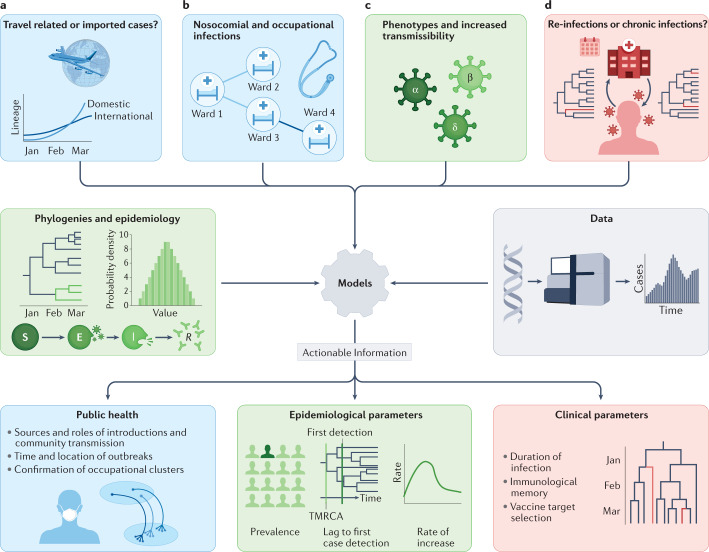
Fig. 1. Phylodynamic approaches to the investigation of SARS-CoV-2 transmission.
Relevant clinical and public health questions are defined (top row), phylodynamic and epidemiological data and models are then combined (middle row), and used in combined or joint analyses to provide actionable insight into virus transmission (bottom row). a | Phylogenetic approaches estimate the rate of international lineage introductions and distinguish introductions from community transmission. b | Genome sequences and phylogenetics support outbreak analyses by identifying or refuting links between local cases; this can lead to identification of outbreak sources and drivers or assessment of nosocomial transmission. c | Phylodynamic techniques using epidemiological demographic models, such as the susceptible–exposed–infected–recovered (SEIR) model, allow us to compare transmission rates between lineages bearing different key genotypes (for example, variants of concern (VOCs) and pre-existing lineages). d | Relative timing of variant and lineage emergence from the global (or regional) phylogeny, and scattering of case genomes across clades can distinguish persistent from repeat infections in some scenarios. Phylogenetics is also useful in studies of lineage turnover and interactions within the host. Panel colours indicate related themes: blue, public health; green, epidemiological parameters; red, clinical parameters. TMRCA, time to the most recent common ancestor.