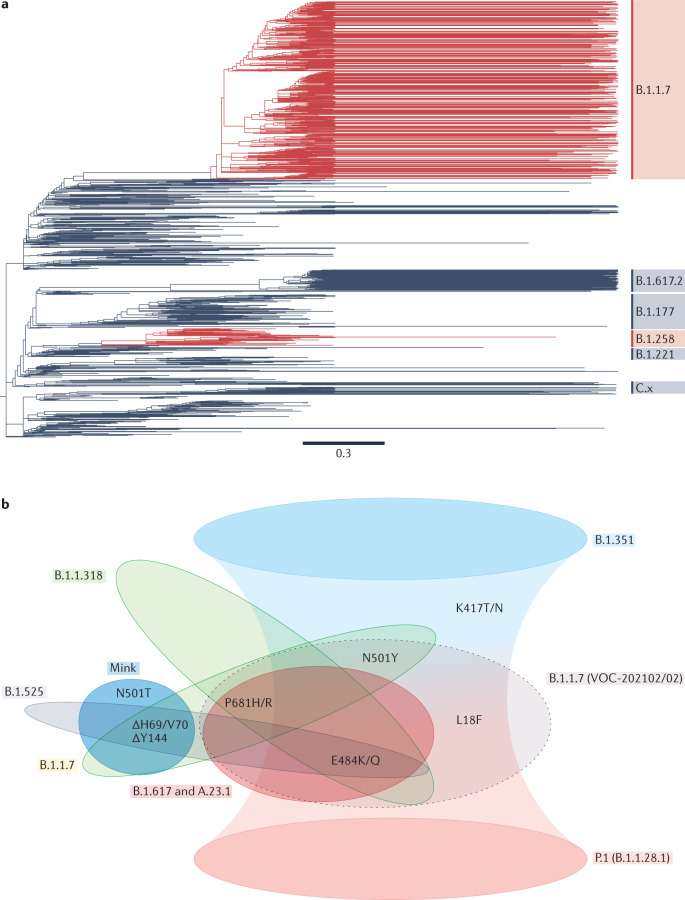
Fig. 3. Convergent evolution of SARS-CoV-2 spike protein.
a | Phylogenies for the first year of the pandemic show the independent emergence of spike ΔH69/V70, indicated in red, in genomes of the B.1.1.7 and B.1.258 lineages respectively — note, the B.1.258 clade in red includes some branches without the deletion. Phylogeny from Nextstrain146,147 (which used data from the Europe ncov GISAID data set148), visualized in Figtree. Acknowledgements of authors responsible for the genetic sequence data generated, shared via the GISAID initiative and used to generate the Nextstrain tree, may be found in Supplementary Table 1. For clarity, not all Pango lineages are shown. b | By the start of 2020 several commonly occurring spike substitutions and deletions had been recognized as shared between lineages. The illustrated substitutions are found in the exposed (that is, outermost on the surface of the virion) subunit of spike, termed S1, or in the spike N-terminal domain (NTD), and are those shared by variants of interest or concern, excluding those shared sporadically or in minor sublineages. B.1.351 and P.1 share K417T/N and (in some B.1.351 sublineages) L18F, as well as two other recurrent substitutions; this is indicated by the overlap of their extended shading. ‘Mink’ refers to the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) mink–human sublineage, termed ‘cluster 5’, which exhibited ΔH69/V70 and N501T (and other spike substitutions)91; the second B.1.1.7 lineage (VOC-202102/02, the grey ellipse with broken-line border) is a cluster of B.1.1.7 that also bears E484K71. N501T is a homoplasy that emerged in mink and may have transferred to humans; it is relatively uncommon, as it was found in only five mink in the original mink farm epidemic in Denmark. Nevertheless, N501T seemed to have emerged independently four times and has been detected in ten human cases149. L18F is an NTD substitution found in some B.1.351 and several of its sublineages, and it is increasing in frequency in B.1.1.7 (ref.67). As in Fig. 2, we see that the same substitutions appear in multiple lineages, implying that they arose independently at different times and places. Here, we also see that not only are individual substitutions shared, but constellations of several changes also seem to co-occur in more than one lineage; this suggests epistatic interactions, with perhaps compensatory changes following immune escape variants.