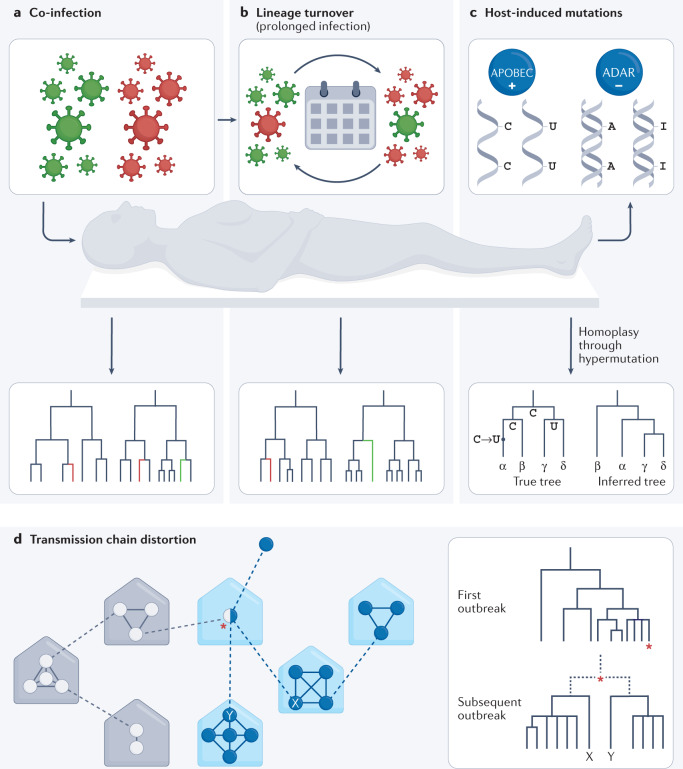
Fig. 4. Effects of within-host evolution and dynamics on epidemiological observations.
Phylogenetic and phylodynamic approaches help detect and understand complex infections, measure within-patient lineage turnover and explore how host-induced mutation affects outbreak investigations. a | Co-infections may confound transmissibility and aetiological studies, but they can be detected using phylogenetics. Specifically, co-infections are identified when viral genomes sequenced from multiple isolates from the same patient are not monophyletic. b | Lineage turnover can occur if within-host lineages share a recent common ancestor and arise from evolution within the host itself. Lineage turnover may complicate patient treatment, as a lineage with lesser susceptibility to host immune responses may give way to a more transmissible lineage after apparently successful completion of a course of therapy. Nevertheless, phylogenetic features, such as longitudinal samples falling into different sister lineages and relative branch lengths, can help detect and account for lineage turnover. c | The antiviral activities of host APOBEC cytidine deaminases, which promote C → U hypermutation, adenosine deaminases that act on RNA (ADARs) and similar host systems, can lead to biases such as C → U homoplasies (convergent evolution) in the case of APOBECs, and changes in virus genome CpG content as a response150–152. Phylogenetics can highlight such convergent changes, which will be seen arising in lineages that are not closely related, and phylogenetic and phylodynamic approaches can be adjusted to account for the elevated rate of particular transitions. d | Co-infections and superinfections can complicate attempts to trace transmission chains, through either lineage turnover or sampling bias (for example, differential PCR amplification or through effects of organotropy). The result can be failure to connect two related transmission chains. A superinfected individual could also cryptically contribute to more than one heterochronous outbreak. The schema shows potential transmission events within households, or similar units (for example, workplaces), in a simplified transmission scenario. The dashed lines indicate transmission events between households. Circles represent individuals, with empty circles indicating infection chains involving lineage 1 and filled circles those involving lineage 2. The red asterisk indicates a co-infected individual who carries both lineages. The phylogeny shows that the true relationship between individuals X and Y may be unclear if lineage 1 dominates the co-infection at the time of sampling.